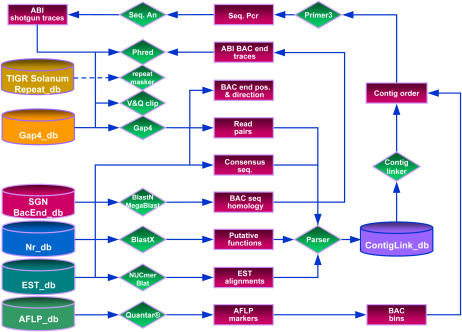
Figure 1.
Schematic overview of the dataflow used in this study. Red-colored rectangles represent datasets, databases are depicted as bins, and applications are drawn as green-colored diamonds. Direction of dataflow is indicated by blue-colored connectors. The dashed blue-colored connector represents an additional step that can be included for repeat masking. For processing raw trace data we rely on PREGAP4 of the Staden package (Bonfield et al., 1995), which is flexible in interfacing a diverse set of tools for base calling, vector clipping, repeat masking, and assembly. In this study we have used PHRED base calling and GAP4 assemblies. From the GAP4 database, consensus sequences and assembly positions are extracted, uploaded, and used by TOPAAS for BLASTX, MUMmer, and BLAT analyses. The system also searches a BAC end database with BLASTN or MegaBlast against consensus sequences. To verify quality, overlap, and direction, corresponding BAC end traces are processed and assembled onto contig sequences. Candidate BAC clones are used for AFLP fingerprint analyses. Comigrating fragments are used to deduce the binning of BACs. Read pair information, BLAST scores, EST alignments, and BAC end positions are parsed into the ContigLinkdb. TOPAAS analyzes the data in ContigLinkdb on a project level and predicts contig links and minimal overlapping BAC clones. BAC binning information is then used for extended contig ordering and selection of minimal overlapping BACs. The primer module part designs nonredundant primers, which are then subsequently used for sequence PCR analysis and gap closure.