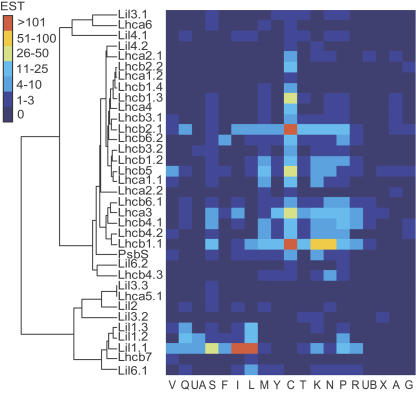
Figure 3.
Digital expression profiling in PopulusDB. Clustered correlation map according to Ewing et al. (1999) showing the distribution of ESTs from the genes in the Lhc supergene family in 19 poplar cDNA libraries; Lil1 genes code for ELIP; Lil2 (OHP1) and Lil6 (OHP2) genes code for OHP; and Lil4 (SEP1) and Lil5 (SEP2) genes code for SEP. Library codes: A, cambial zone (A + B); C, young leaves; F, flower buds; G, tension wood; I, senescing leaves; UA, active cambium; UB, dormant cambium; K, apical shoot; L, cold-stressed leaves; M, female catkins; N, bark; P, petioles; Q, dormant buds; R, roots; S, imbibed seeds; T, shoot meristem; V, male catkins; X, wood cell death; Y, virus/fungus-infected leaves; for a detailed library description, see Sterky et al. (2004) or www.populus.db.umu.se.