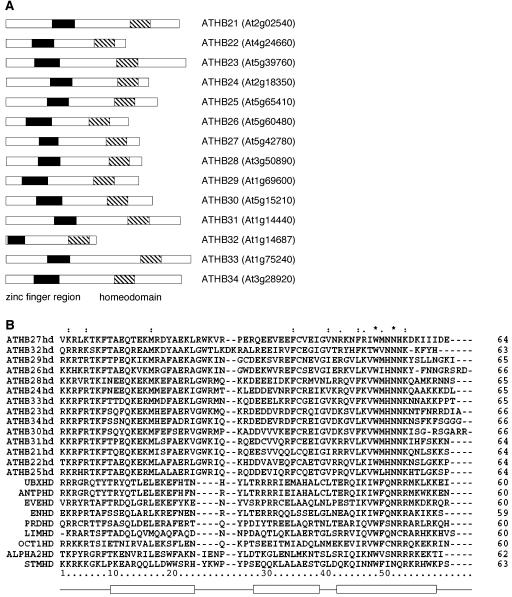
Figure 1.
The Arabidopsis ZF-HD family. A, The 14 members of the Arabidopsis ZF-HD family are shown schematically, and their corresponding Arabidopsis Genome Initiative locus identifiers listed. The ZF region is shown as a black box, and the HD is represented as a striped box. B, The HD's of the Arabidopsis ZF-HD family members, along with other representative HD-containing proteins are aligned, including Arabidopsis SHOOT MERISTEMLESS (STMHD; At1g62360/Q38874; Long et al., 1996), M. musculus L3 (LIMHD; BAA21649; Matsumoto et al., 1996), as well as the HDs which have been crystallized: D. melanogaster Antennapedia (ANTPHD; CAA43307; Fraenkel and Pabo, 1998), D. melanogaster Even-skipped (EVEHD; AAA28522; Hirsch and Aggarwal, 1995), D. melanogaster Engrailed (ENHD; AAA65478; Kissinger et al., 1990), D. melanogaster Paired (PRD; P06601; Wilson et al., 1995), yeast MATα2 (ALPHA2HD; Q6B2C0; Wolberger et al., 1991), Xenopus laevis Oct-1 (OCT1HD; CAA35051; Klemm et al., 1994), and D. melanogaster Ubx (UBXHD; CAA53803; Passner et al., 1999). Positions of helices 1, 2, and 3 are shown below. Asterisk (*) indicates positions which have a single, fully conserved residue; Colon (:) indicates that one of the following strong groups is fully conserved: STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, and FYW; Period (.) indicates that one of the following weaker groups is fully conserved: CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, and HFY. These are all the positively scoring groups that occur in the Gonnet Pam250 matrix. The strong and weak groups are defined as strong score >0.5 and weak score <0.5, respectively.