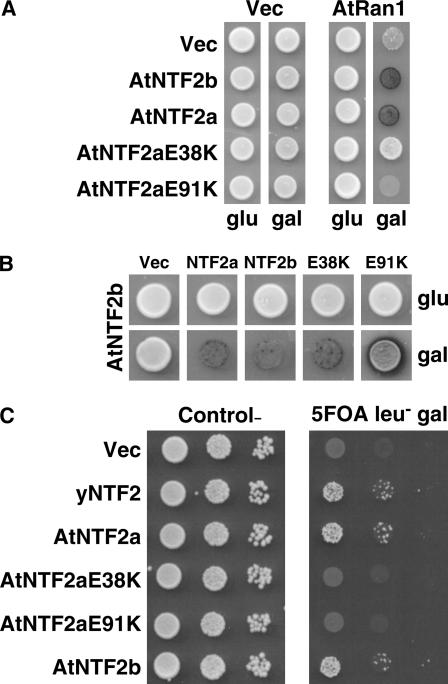
Figure 4.
E38K and E91K mutations in AtNTF2a disrupt Ran binding and AtNTF2a function in vivo. A, Yeast two hybrid was used to assay the interaction between wild-type and mutant AtNTF2a and Arabidopsis Ran (AtRAN1) or a vector control (Vec). Neither AtNTF2aE38K nor AtNTF2aE91K interacted with AtRAN, as indicated by loss of the blue color indicative of β-galactosidase expression in these spots. As controls, wild-type AtNTF2a and AtNTF2b both interacted with AtRAN in this experiment. No interaction was observed with the vector control. B, AtNTF2a mutants interact with AtNTF2b. The interaction of AtNTF2b with itself and with both wild-type and mutant AtNTF2a was examined by two-hybrid assay. The blue color indicative of β-galactosidase expression reveals that AtNTF2b interacts with all versions of NTF1/2 tested, indicating that the mutant AtNTF2a proteins are expressed and capable of interacting in the two-hybrid assay. C, The function of AtNTF2aE38K and AtNTF2aE91K was examined by a plasmid shuffle assay. Yeast cells deleted for the genomic copy of the essential NTF2 gene but maintained by an NTF2 genomic plasmid (see “Materials and Methods”) were transformed with plasmids expressing AtNTF2a, AtNTF2aE38K, AtNTF2aE91K, or as controls, vector, yNTF2, or AtNTF2b under the control of a Gal-inducible promoter. Cultures were grown to saturation, serially diluted, and spotted on control (left) or 5-FOA (right) plates to eliminate the NTF2 maintenance plasmid. Only wild-type AtNTF2a and AtNTF2b supported cell growth, as did the control (yNTF2).