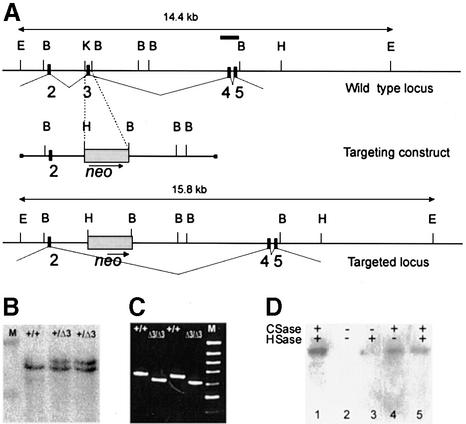
Fig. 1. Deletion of exon 3 in the perlecan (Hspg2) gene. (A) Region of the perlecan gene containing exons 2–5 (top), the targeting construct (middle) and the region after homologous recombination. Exons are depicted as black boxes and numbered. A 130 bp KpnI–BamHI fragment containing exon 3 was replaced by the PGK-neo selection cassette. The probe containing exons 4 and 5 that was used for Southern blots is depicted as a bar (top). E, EcoRI; B, BamHI; H, HindIII; K, KpnI. (B) Southern blot analysis of EcoRI-digested DNA from ES cell clones. M, λ DNA digested with HindIII, upper band 19.9 kb, lower band 9.5 kb. (C) Products of RT–PCR reactions performed with primers coding for the N-terminal region of the perlecan protein (for sequences, see text) separated in an agarose gel. Sources of RNA were whole E16 embryos and primary fibroblasts isolated from newborn wild-type(+/+) or Hspg2Δ3/Δ3 mice. M, 100 bp ladder, uppermost band 1000 bp. The PCR products are 650 bp in wild-type and 605 bp in mutant samples. (D) Immunoblot analysis of proteoglycan-enriched fractions isolated from the culture medium of wild-type and Hspg2Δ3/Δ3 fibroblasts. Samples were treated with (+) or without (–) chondroitinase (CSase) and heparitinase (HSase), subjected to SDS–PAGE in a 5% gel, and resolved by immunoblotting with an antibody against perlecan. Lane 1, wild-type sample treated with chondroitinase and heparitinase; lane 2, untreated mutant sample; lane 3, mutant sample treated with hepari tinase only; lane 4, mutant sample treated with chondroitinase only; lane 5, mutant sample treated with chondroitinase and heparitinase. The perlecan core protein produced by the mutant cells was of the same size as the wild-type perlecan, and core protein was observed after chondroitinase treatment, indicating that the major portion of the mutant perlecan carries chondroitin sulfate chains only.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.