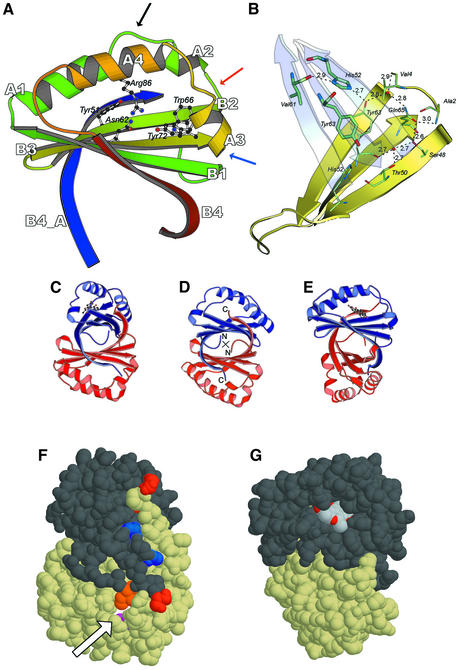
Fig. 2. Structural model of ActVA-Orf6 monooxygenase. (A) The B subunit of ActVA-Orf6 monooxygenase viewed from the active site entrance is characterized by a ferredoxin fold, with four β-strands (B1–B4) and four α-helices (A1–A4). The blue strand is the C-terminal domain swap contributed by the A subunit. The mobile loop 34–38 (black arrow), the type II β-turn (red arrow) and the type III β-turn (blue arrow) are highlighted. Residues involved in the enzymatic mechanism are drawn as ball-and-stick. (B) The monomer–monomer interface. The interaction between the two β-sheets (yellow and cyan) is stabilized through a network of inter- and intramolecular hydrogen bonds, which also restrain the N-terminus (residues 2 and 4). Thick and thin sticks represent residues contributed, respectively, by the two subunits. (C) The homodimer of ActVA-Orf6 with nanaomycin D bound to subunit A, in the same orientation as in (F). (D) The homodimer of ActVA-Orf6. The 2-fold axis of symmetry is perpendicular to the plane of the picture (cross). (E) The homodimer of ActVA-Orf6 with nanaomycin D bound to subunit A, in the same orientation as in (G). (F) The bottom of the ActVA-Orf6 active site in the B subunit (yellow van der Waals spheres) shows a narrow tunnel (arrow) gated by Gln37 (purple). Ile110 (orange), belonging to the C-terminal swap of the other monomer, also contributes to the closure of the back of the active site. C- (red) and N- (blue) termini are also shown. (G) The dimer in a different orientation, showing nanaomycin D bound to subunit A (dark grey van der Waals spheres) of ActVA-Orf6 monooxygenase and exposing part of the pyrano-γ-lactone ring to the solvent. This moiety (and C-15 methyl, in particular) obstructs the active site entrance after binding.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.