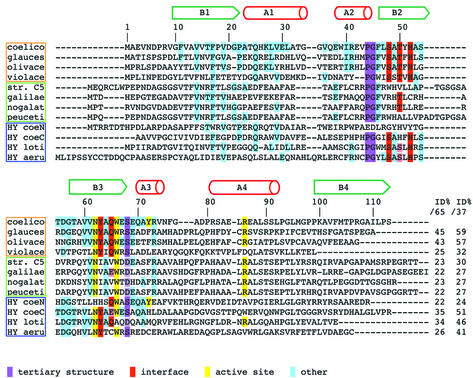
Fig. 3. Multiple sequence alignment of proteins similar to ActVA-Orf6 monooxygenase. The sequences have been selected from the output of a PSI-BLAST search performed using ActVA-Orf6 monooxygenase as query. Not all hits are shown in the alignment, and the alignments are less reliable from residue 75. Above the alignment, the secondary structure of ActVA-Orf6 is shown. The genes are clustered in three groups: a family of monooxygenases with an interface identical to ActVA-Orf6 monooxygenase (orange frame); a second family of putative Streptomyces monooxygenases that probably possess a different interface (green frame); and hypothetical proteins of unassigned function (blue frame). Aligned sequences: protein, organism (gene name and accession No. in SWALL database in parentheses): coelico, ActVA-Orf6 monooxygenase, S.coelicolor (ActVA-Orf6, Q53908); glauces, TcmF1 monooxygenase, S.glaucescens; (TcmH, P39889); olivace, TcmF1 monooxygenase, S.olivaceus (ElmH, Q9L4Y0); violace, C-5 anthrone oxidase, S.violaceus (Jad-Orf7, Q56157); str. C5, oxygenase, unspeciated streptomycete strain C5 (Streptomyces sp. C5; from the Frederick Cancer Research Council) (DauA-OrfE, Q55222); galilae, aklanonic acid anthrone monooxygenase, S.galilaeus (AknX, Q9L552); nogalat, SnoB protein, S.nogalater (SnoB, Q54493); peuceti, anthraquinol monooxygenase, S.peucetius (Dps-Orf8, Q54813); HY coeN, N-terminus of hypothetical 26.4 kDa protein, S.coelicolor (SCI7.27C, Q9X9W3); HY coeC, C-terminus of the same 26.4 kDa protein, S.coelicolor; HY loti, hypothetical protein, Mesorhizobium loti (MLR8211, Q983R7); HY aeru, hypothetical protein, Psuedomonas aeruginosa (PA2274, Q9I1K0). The biochemical function has been assigned experimentally for ActVA-Orf6 monooxygenase from S.coelicolor, TcmH from S.glaucescens, ElmH from S.olivaceus and AknX from S.galilaeus. For all the other proteins, the function has been deduced on the basis of homology with characterized proteins, although other indirect evidence may be available. Full boxes highlight conserved residues: purple, residues essential for tertiary structure (β-turns); red, interface polar residues (quaternary structure); yellow, active site residues; cyan, other identities. Similarities are only reported if they are consistent with structural data: light purple (tertiary structure) and light red (interface). End of the alignment: the first column reports percentage identity calculated for the ‘ferredoxin unit’ lacking helix A4 and the N-terminus (ID%/65, residues 10–74 of ActVA-Orf6 monooxygenase); the second column refers to the ‘minimal structural/functional unit’ (ID%/37, residues 38–74).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.