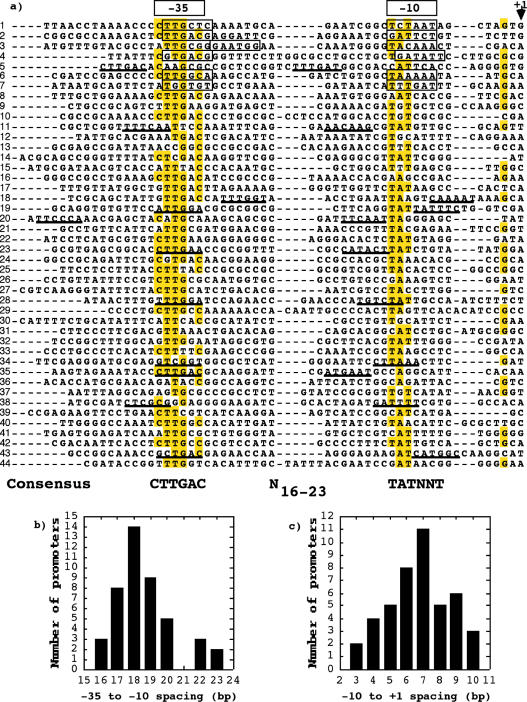
Figure 1.
(a) Alignment of the promoter regions identified in R.etli. The position of the −10 and −35 elements are indicated at the top of the alignment. The −10 and −35 elements identified by other authors are indicated within boxes. The nucleotides that appear in ≥51% of the cases in any given position are marked in yellow. The transcription start-sites are marked with +1. Alternative promoters with suboptimal properties are underline. The consensus sequence derived from the alignment are indicated at the bottom of the alignment. The promoters identified were: 1, prepA; 2, pctRNA; 3, precA; 4, plipA; 5, pthiC; 6, pnifR promoter 1; 7, pnifR promoter 2; 8, psigA; 9, pypf00079; 10, pglyQ; 11, pyhch00468; 12, pyhch00334; 13, pyhch01154; 14, peda; 15, pyhch00528; 16, pengB; 17, p16S rRNA; 18, pyhch01141; 19, pyhch00226; 20, pyhch00739 pseudogene; 21, phemC; 22, pclpP; 23, prpoH1; 24, paroG; 25, pypch00196; 26, pureA; 27, pypch00131; 28, pndvB; 29, pype00047 pseudogene; 30, pyhch00197; 31, ppfs; 32, pcycH; 33, pypch00552; 34, pyhch00096; 35, pypch00132; 36, ppepN promoter 1; 37, ppepN promoter 2; 38, pyhch00326; 39, prpsP; 40, pyhch01115; 41, pyhch00280; 42, pispB; 43, pyhch00076; 44, pexoR2. (b) Distribution of spacer lengths for the 44 promoters of the (a) (c) Distribution of the distance between the 3′ of the −10 element (TATNNT) and the +1