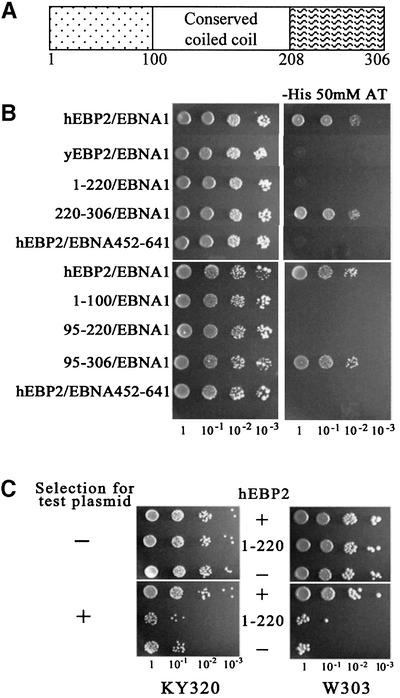
Fig. 6. The EBNA1 binding region of hEBP2 is required for EBNA1-mediated segregation. (A) Schematic representation of hEBP2 showing the most conserved region, which corresponds to a predicted coiled-coil domain. Amino acids numbers are indicated. (B) EBP2 from yeast (yEBP2) and humans (hEBP2), and hEBP2 fragments 1–220, 220–306, 1–100, 95–220 and 95–306 were tested for binding to EBNA1 in a yeast two-hybrid assay, as determined by activation of the HIS3 reporter gene. As a negative control, hEBP2 was tested for binding to EBNA452–641, which lacks the hEBP2 binding sequence. Ten-fold serial dilutions of log-phase cultures were grown on plates containing histidine (left panel) or lacking histidine and containing 50 mM AT (right panel). (C) Plasmid loss assay comparing the ability of hEBP2 (+) and the hEBP2 1–220 mutant (1–220) to support the segregation of YRp7FR in the presence of EBNA1. Assay results in the absence of hEBP2 expression (–) are also shown. Assays were conducted as in Figure 3.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.