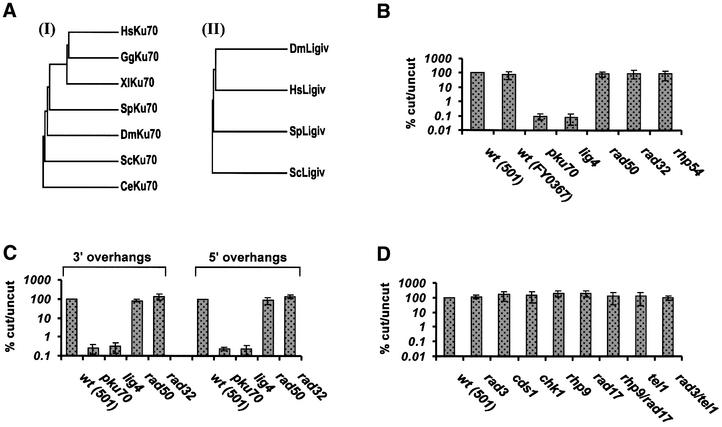
Fig. 1. Genetic requirements for NHEJ in S.pombe. (A) Phylogenetic trees showing the evolutionary relationship of (I) Ku70 and (II) DNA ligase IV proteins from different organisms. The trees are based on multiple sequence alignments created using the PILEUP program of the GCG software. DDBJ/EMBL/GenBank accession Nos are as follows: HsKu70 (Homo sapiens), P12956; GgKu70 (Gallus gallus), O93257; XlKu70 (Xenopus laevis), BAA76953; SpKu70 (Schizosaccharomyces pombe), CAA22471; DmKu70 (Drosophila melanogaster), Q23976; ScKu70 (Saccharomyces cerevisiae), P32807; CeKu70 (Caenorhabditis elegans), CAB55094; DmLigIV (D.melanogaster), AAF48298; HsLigIV (H.sapiens), NP_002303 with the addition of 67 amino acids at the N-terminus as described in Grawunder et al. (1998); SpLigIV (S.pombe), CAA21085; ScLigIV (S.cerevisiae), Q08387. (B–D) Analysis of NHEJ in S.pombe mutants using a plasmid-based assay: (B and D) blunt DSBs, (C) cohesive DSBs. FY0367 is the pku70 parental strain.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.