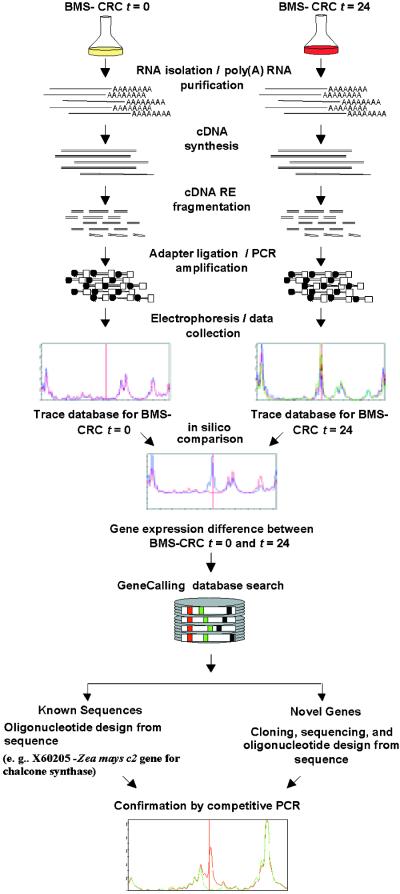
Figure 1.
Schematic of the GeneCalling Analysis.
Total RNA is extracted from the cell lines, and poly(A)+ RNA is purified and converted to cDNA, which is fragmented by using pairs of restriction enzymes (RE). Adapters are then ligated to the ends of the fragments, which are amplified by polymerase chain reaction (PCR). Because one of the PCR primers is labeled with a fluorescent tag (fluorescamine; FAM), amplified fragments can be visualized during electrophoresis. For each sample and restriction enzyme pair, electronic images of the gel lane traces are collected and kept in a sample trace database. Comparisons of the trace databases reveal specific expression differences that are characterized by the length of the amplified fragment and by restriction enzyme sequence information. The identity of each differentially expressed gene fragment can be established either by performing a GeneCalling search in a sequence database or by cloning and sequencing the desired cDNA fragment. The identity of the cDNA fragment is confirmed by competitive PCR in which the original PCR reaction is reamplified in the presence or absence of an excess of an unlabeled, gene-specific PCR primer. Further characterization of known and newly discovered sequences that are identified by GeneCalling analysis as differentially expressed can be obtained by BLASTX and BLASTN analyses against public and proprietary databases. t, time in hours.