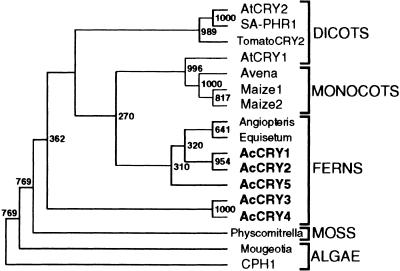
Figure 8.
Phylogenetic Analysis of Plant Cryptochromes.
The regions of partial amino acid sequences used to generate the phylogenetic tree are shown in Figure 1. The neighbor-joining method was used to calculate bootstrap values from 1000 replicates (http://www.genome.ad.jp/) and to draw the tree by using TreeView software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The numbers located at branch points are the bootstrap values. GenBank/DDBJ/EMBL accession numbers for each cryptochrome DNA sequence, except for those given in the legend of Figure 1, are as follows: Angiopteris for Angiopteris evecta, X99261; Avena for Avena sativa, X99262; CPH1 for Chlamydomonas CPH1, L07561; Equisetum for Equisetum arvense, X99263; Maize1 and Maize2 for Zea mays, X99265 and X99266; Mougeotia for Mougeotia scalaris, AJ000694; Physcomitrella for P. patens, AB027528; SA-PHR1 for S. alba SA-PHR1, X72019; and TomatoCRY2 for Lycopersicon esculentum, AJ000695.