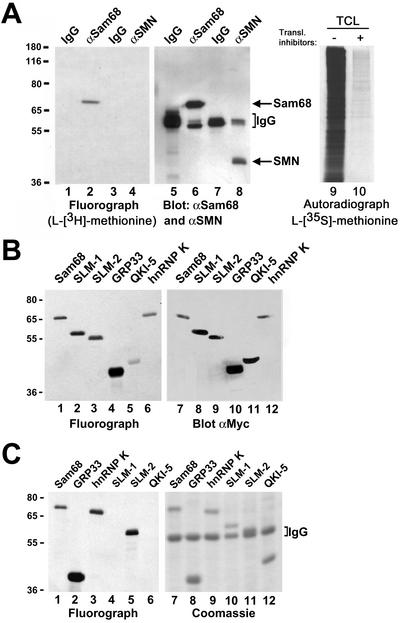
Figure 1.
In vivo methylation of Sam68 and other KH domain RNA binding proteins. (A) In vivo methylation labeling was performed by metabolically labeling cells with [l-[methyl-3H]methionine for 3 h in the presence of cycloheximide and chloramphenicol. The cell lysates were immunoprecipitated with anti-Sam68 (mAb 7-1) and anti-SMN antibodies. The 3H-labeled proteins were separated by SDS-PAGE and visualized by fluorography (lane 1–4; exp. 24 h), and the proteins were visualized by immunoblotting with anti-Sam68 and anti-SMN antibodies (lanes 5–8). The migration of Sam68 and SMN is shown. The heavy chain of IgG is indicated with a bracket. TCLs of HeLa cells metabolically labeled with l-[35S]methionine in the absence (lane 9; −) or presence (lane 10; +) of protein synthesis inhibitors were separated by SDS-PAGE and visualized by autoradiography (exp. 24 h). (B) Metabolic labeling for the in vivo methylation was performed 24 h after transfection of myc-epitope–tagged Sam68, SLM-1, SLM-2, GRP33, QKI-5, and hnRNP K expression vectors in HeLa cells. The cell lysates were immunoprecipitated with anti-myc antibodies and the bound 3H-labeled proteins were separated by SDS-PAGE and visualized either by fluorography (lanes 1–6; exp. 12 h) or by immunoblotting with the anti-myc antibody (lanes 7–12). (C) Endogenous protein methyltransferase activity coimmunoprecipitates with KH domain-containing proteins. HeLa cells were transfected with myc-epitope–tagged Sam68, SLM-1, SLM-2, GRP33, QKI-5, and hnRNP K. The cells were lyzed and myc immunoprecipitations were incubated with [3H-methyl]S-adenosyl-l-methionine in vitro. The immunoprecipitated proteins were visualized by Coomassie staining (lanes 7–12) or by fluorography (lanes 1–6; exp. 6 h). The migration of the heavy chain of IgG is indicated.