Figure 6.
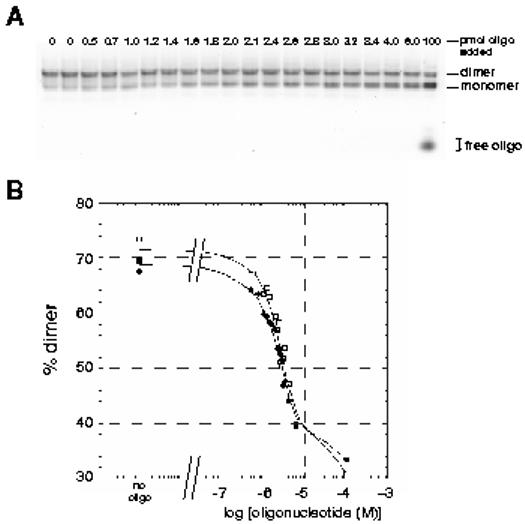
Effect of challenging pre-formed HIV-2 RNA dimers with increasing concentrations of oligonucleotide asRODPBS and asROD309. A) Increasing amounts of oligonucleotide asRODPBS were added to pre-formed dimers of RNA 1-381 and incubated for an additional 37°C for 15 minutes. One microliter aliquots of oligonucleotide stock solutions were added to five picomoles of RNA in a final reaction volume of 10μl (final oligonucleotide concentrations ranged between 0 and 10 μM). Reactions were loaded onto TBM agarose gels and electrophoresis was carried out at 4°C. Ethidium bromide staining was used to visualize the monomer and dimer RNA bands. B) Quantification of dimer yield from the gel shown in panel A and for the same experiment using oligonucleotide asROD309. A Fuji FLA-3000G Fluorescence Imager and Image Gauge software were used to determine the fraction of RNA in the dimer band of each lane. These values were plotted versus the log of the oligonucleotide concentration for the two oligonucleotides found to be effective inhibitors of dimerization, namely asRODPBS (open squares) and asROD309 (closed squares). Other oligonucleotides tested did not effectively inhibit dimerization in this assay, even at high concentrations.
