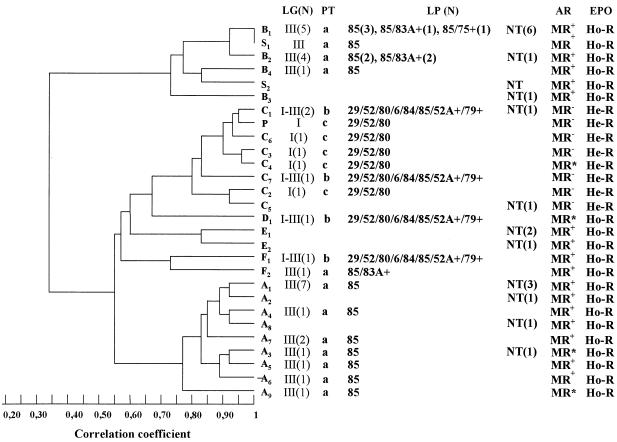
FIG. 1.
(Left) Dendrogram of the similarity between SmaI-digested DNA patterns obtained by PFGE from representative MRSA strains of each pulsotype and subtype isolated in Córdoba, Argentina, and representative South American (S1 and S2) and pediatric (P) clones described in Buenos Aires, Argentina. (Right) Distribution of the lytic groups (LG), phage types (PT), lytic phages (LP), antibiotic resistance pattern (AR), and phenotypic evaluation of methicillin resistance (EPO) of each subtype of MRSA clinical isolates from Córdoba, Argentina. NT, nontypeable; N, number of isolates; MR+, strains resistant to β-lactam antibiotics, ciprofloxacin (CIP), clindamycin (CLI), erythromycin (ERY), and gentamicin (GEN) and variably resistant to minocycline, chloramphenicol (CHL), trimethoprim-sulfamethoxazole, and rifampin (RIF); MR−, strains resistant only to β-lactam antibiotics and GEN; MR*, strains resistant to β-lactam antibiotics and GEN and variably resistant to RIF, ERY, CLI, CIP, and CHL; Ho-R, homoresistant strains; He-R, heteroresistant strains.