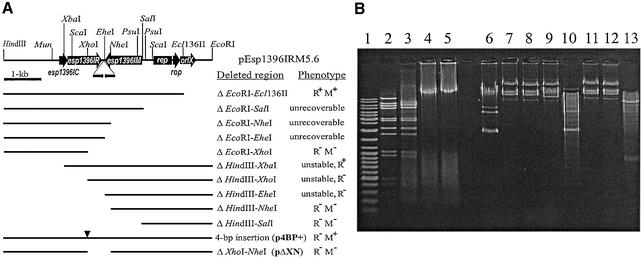
Figure 1.
(A) Schematic map of pEsp1396IRM5.6 plasmid and deletion mapping of genes encoding the Esp1396I RM system. Thin line represents the entire pEsp1396 plasmid cleaved with BglII and inserted into pUC19, which was opened with BamHI. Targets for both HindIII and EcoRI are from pUC19 multiple cloning site. The identified ORFs are shown in accordance with the sequencing data. Directions of transcription of all identified genes are indicated by arrows. Deletion plasmids constructed in this work are indicated below as thin lines and their relevant phenotypes are indicated on the right. Triangle marks the 4-bp insertion introduced at the XhoI site in esp1396IR and resulting in p4BP+. Restriction and modification phenotypes were determined as described under Materials and Methods. M+, Mtase protection from Esp1396I cleavage; M–, no Mtase protection from Esp1396I cleavage; R+, Enase activity; R–, Enase activity not detectable. (B) Restriction and modification activities of cloned genes. Lane 1, DNA ladder; lane 2, λ DNA incubated with Esp1396I; lanes 3–5, λ DNA incubated with crude extracts prepared from cells carrying either pEsp1396IRM5.6 or derivatives p4BP+ and pΔXN, respectively; lanes 6–9, DNAs isolated from cells carrying pUC19, pEsp1396IRM5.6, p4BP+ and pΔXN, respectively; lanes 10–13, the same as in lanes 6–9 but after the incubation with Esp1396I.