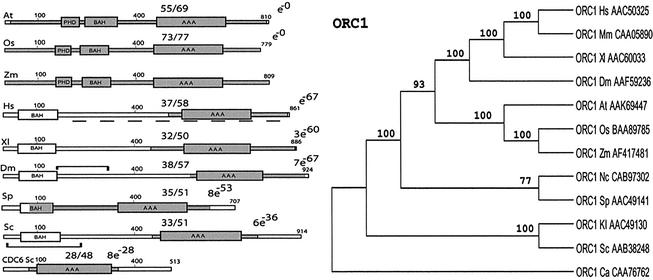
Figure 1.
Modular architecture of ORC1 homologs and the phylogenetic tree. Proteins are drawn to scale, the figures along the bars reflect amino acid numbers. The numbers separated by a solidus show the degree of identity/similarity between the maize proteins and the other ORC subunits. Regions displaying similarity to the maize protein are shaded. The boxes correspond to the architectural motifs. The brackets in the DmORC1 and ScORC1 molecules indicate the regions binding HP1 and SIR1, respectively. The broken line under the human ORC1 corresponds to the domain interacting with HBO1. In the NJ tree branch lengths are proportional to amino acid substitutions (bar represents 10 substitutions per 100 residues). Bootstrap values (500 pseudoreplicates) are shown in association with nodes in the phylogeny. Branches with bootstrap values <50% are collapsed. Abbreviations: Sc, S.cervisiae; Sp, S.pombe; Dm, Drosophila melanogaster; At, Arabidopsis thaliana; Zm, Zea mays; Os, Oryza sativa; Xl, X.laevis; Hs, Homo sapiens; Mm, Mus musculus; Kl, K.lactis; Nc, N.crassa; Ca, Candida albicans.