Figure 1.
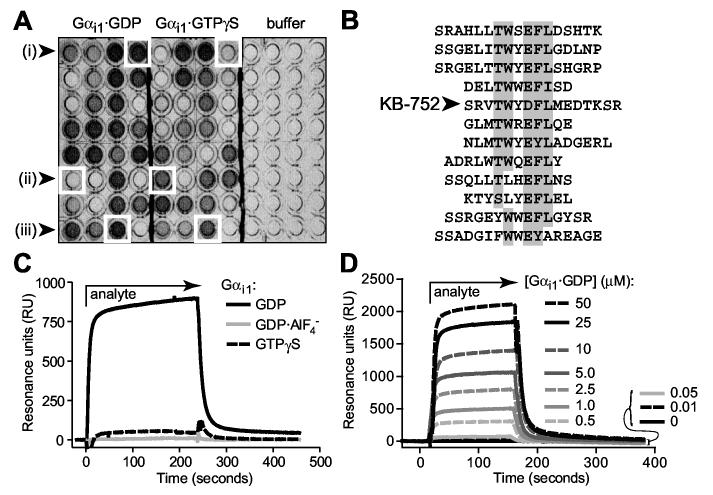
Identification of nucleotide-dependent Gαi-binding peptides via phage display. (A) Representative phage ELISA results indicating the identification of (i) GDP-selective, (ii) GTPγS-selective, and (iii) nucleotide state-independent Gαi1-binding peptides. (B) Sequences of 12 isolated peptides with GDP-selective binding to Gαi1, sharing a consensus TWXE/DFL motif with the particular peptide used in this study: KB-752. (C) Nucleotide-dependent binding of KB-752 as measured by surface plasmon resonance (SPR). 5 μM Gαi1 protein (“analyte”), in each of three nucleotide bound states, was injected over immobilized, biotinylated KB-752. Non-specific binding to a control peptide was subtracted from each curve. (D) GDP-bound Gαi1 was injected at each indicated concentration over immobilized KB-752 to determine the dissociation constant (Kd) for this interaction pair. SPR-derived dissociation constants for the interaction of KB-752 with Gαi1 and Gαo, in their ground state (GDP-bound), transition state-mimetic (GDP·AlF4- bound), and activated state (GTPγS-bound) forms, were obtained from analyses (n = 4-6 for each state) similar to that shown in panel D. Dissociation constants of >1000 μM were obtained for both Gα subunits in their GDP·AlF4--bound form, and for Gαo bound to GTPγS.
