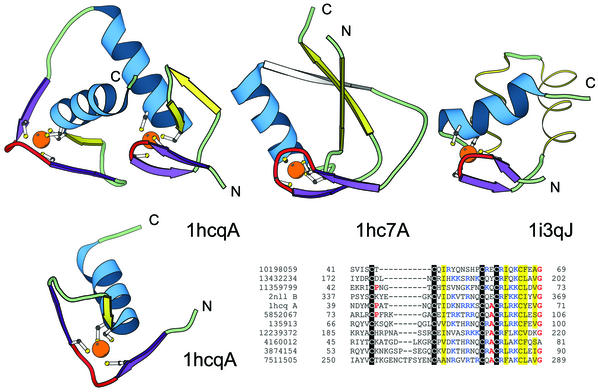
Figure 4.
Structural comparisons of representative treble clef fingers. Structural diagrams of the treble clef of the steroid hormone ‘estrogen’ receptor DNA-binding domain showing both zinc-binding sites (1hcq, chain A, residues 3–36 and 39–71), prolyl-tRNA synthetase (1hc7, chain A, residues 454–477; 418–440) and the RPB10 protein from RNA polymerase II (1i3q, chain J, residues 3–54) are shown to illustrate the wide range of variations in the structure of the treble clef finger. The N-terminal site of 1hcq is a typical treble clef motif. The C-terminal site (below), reoriented for clarity, lacks the β-hairpin shown in yellow for the N-terminal treble clef and has a distorted knuckle (red). This zinc-binding region shows some resemblance to other treble clef fingers in the placement of the ligands for zinc binding. However, the knuckle, which contributes the other two ligands in treble clef fingers, is significantly different and does not align structurally with those in other fingers. PSI-BLAST (33,34) alignment of representative nuclear receptor sequences. The sequences included in the alignment are: the nuclear factor NHR-63 (gi 10198059), peroxisome proliferator activated receptor (gi 13432234), NHR-18 (gi 11359799), retenoid X receptor (2nll), estrogen receptor (1hcq), tailless protein (TLL_DROME gi 135913), FTZ-F1 (gi 12239372), dissatisfaction (gi 4160012), NHR-67 (gi 3874154) and NHR-2 (gi 7511505). The gi accession number, start and end sequence number are given. The coloring scheme follows Figure 1.