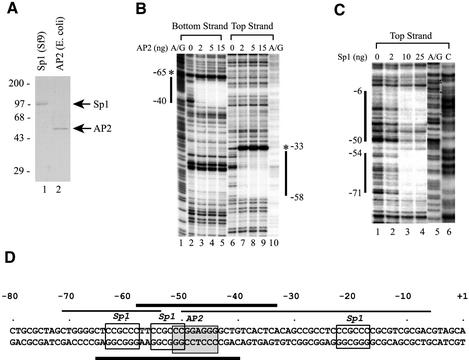
Figure 3.
Sp1 and AP2 proteins bind directly to their cognate sites in the hTAFII55 promoter-proximal region. (A) Purified recombinant Sp1 and AP2 proteins. FLAG-tagged Sp1 and FLAG-tagged AP2 proteins were purified from insect cells and bacteria, respectively, as described in Materials and Methods. The proteins were then loaded on a 10% polyacrylamide–SDS gel and visualized after Coomassie blue staining. Positions of prestained protein size markers (in kDa) are indicated on the left. (B) DNase I footprinting with recombinant AP2 protein. End-labeled top and bottom strands of the hTAFII55 promoter fragment spanning –128 to +87 were incubated, respectively, with different amounts of FLAG-tagged AP2 protein as indicated. DNase I footprinting was then performed as described in Materials and Methods, and visualized after exposure to an X-ray film. The A/G footprinting markers generated by the Maxam–Gilbert sequencing method (85) were included to determine the boundaries of the protected regions (marked by vertical lines). Hypersensitive sites induced by AP2 binding are denoted by asterisks. (C) DNase I footprinting with recombinant Sp1 protein. DNase I footprinting with FLAG-tagged Sp1 was similarly performed as described above, except using only the top strand of the labeled hTAFII55 fragment and including an additional footprinting marker ‘C’ generated by the Maxam–Gilbert sequencing method. (D) Summary of the hTAFII55 promoter region protected by Sp1 and AP2. The hTAFII55 promoter sequence spanning –80 to +1 were illustrated with core sequences recognized by Sp1 and AP2 indicated by boxes. Horizontal lines mark the sequences protected by AP2 (thick bars) and Sp1 (thin bars) proteins in DNase I footprinting experiments.