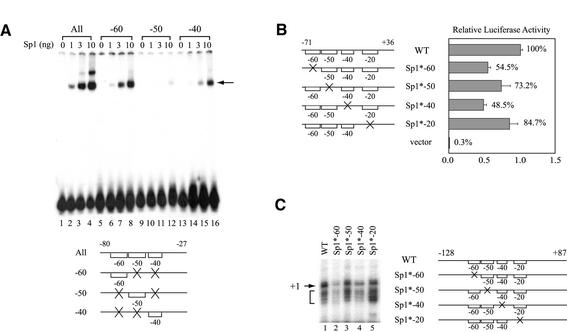
Figure 5.
Sp1-binding sites located at the hTAFII55 promoter-proximal region are functionally involved in hTAFII55 gene expression. (A) Relative binding affinities of the three upstream Sp1-binding sites. The EMSA was performed with different amounts of FLAG-tagged Sp1 protein using equal amounts (6 fmol) of 32P-labeled hTAFII55 promoter fragments as indicated at the bottom. Brackets represent intact binding sites and ‘X’ denotes mutations at the motif. The Sp1–DNA complex is indicated by an arrow. (B) Reporter gene assay with promoter constructs containing individually mutated Sp1-binding sites. Transient transfection was performed in C-33A cells using reporter constructs pGL2-TAF55(–71/+36), pGL2-TAF55(–71/+36)Sp1*-60, pGL2-TAF55(–71/+36)AP2*, pGL2-TAF55(–71/+36)Sp1*-40, pGL2-TAF55(–71/+36)Sp1*-20 and pGL2-Basic, respectively. Reporter gene activities were normalized to that of the wild-type construct. (C) In vitro transcription assay with promoter constructs containing individually mutated Sp1-binding sites. Transcription reactions were performed in HeLa nuclear extracts with hTAFII55 promoter constructs pGL2-TAF55(–128/+87), pGL2-TAF55(–128/+87)Sp1*-60, pGL2-TAF55(–128/+87)AP2*, pGL2-TAF55(–128/+87)Sp1*-40 and pGL2-TAF55(–128/+87)Sp1*-20, which contain mutations at each of the Sp1-binding sites. The amounts of RNA synthesized were quantified by primer extension using Luc(AS)-1 primer (55). The arrow points to the major transcription start site (+1) and the bracket denotes minor start sites. Promoter constructs used in this experiment are illustrated on the right with brackets representing intact Sp1 sites, and an ‘X’ indicating a mutated site.