Abstract
We have developed a functional genomics tool to identify the subset of cDNAs encoding secreted and membrane-bound proteins within a library (the ‘secretome’). A Sindbis virus replicon was engineered such that the envelope protein precursor no longer enters the secretory pathway. cDNA fragments were fused to the mutant precursor and expression screened for their ability to restore membrane localization of envelope proteins. In this way, recombinant replicons were released within infectious viral particles only if the cDNA fragment they contain encodes a secretory signal. By using engineered viral replicons to selectively export cDNAs of interest in the culture medium, the methodology reported here efficiently filters genetic information in mammalian cells without the need to select individual clones. This adaptation of the ‘signal trap’ strategy is highly sensitive (1/200 000) and efficient. Indeed, of the 2546 inserts that were retrieved after screening various libraries, more than 97% contained a putative signal peptide. These 2473 clones encoded 419 unique cDNAs, of which 77% were previously annotated. Of the 94 cDNAs encoding proteins of unknown function, 24% either had no match in databases or contained a secretory signal that could not be predicted from electronic data.
INTRODUCTION
Secreted and membrane-bound proteins are key players in various biological processes: homeostasis, hormonal regulation, cell adhesion, intercellular communication, etc. The vast majority of these proteins are synthesized as precursors that are characterized by a short N-terminal hydrophobic sequence. This ‘signal peptide’ serves to direct the nascent polypeptide chain to the cellular secretory pathway and to mediate its translocation across the lipid bilayer (1). Because of their therapeutic potential, great efforts are deployed to identify genes encoding secreted and membrane-bound proteins. Selective retrieval of cDNAs encoding secreted proteins can be performed according to the ‘signal trap’ methodology (2). This consists of transfecting a library of cDNA fragments fused to a reporter and isolating cells harboring an extracellular reporter. The presence of the reporter in the extracellular space is indicative that the cDNA fragment encodes a functional signal peptide. Typically, reporters are proteins that are normally secreted or membrane-bound but that have been engineered such that the function of their native signal peptide is abolished. Examples of reporters used in conventional signal traps include mutated forms of yeast invertase (3), IL-3 receptor (4) or alkaline phosphatase (5). Whatever the reporter that is used, signal traps currently involve the selection of transfected cells displaying an active extracellular reporter. The need to select cells greatly limits the usefulness and the throughput of the method because selection is achieved either after limiting dilution, by colony picking or using sophisticated sorting equipment. In addition, the cell selection procedure may increase the occurrence of false positives. In fact, the cell selection step is merely a prerequisite to obtain the nucleic acid inserted in the expression vector through PCR, plasmid rescue or related techniques (6).
We have therefore sought to develop an expression screening technology in which activation of the reporter triggers the ‘export’ of the target nucleic acid out of the cells into the culture medium, thereby obviating the need to select cells. This was done by exploiting the natural property of viruses to export genetic material out of cells. More specifically, the Sindbis virus genome was engineered such that the envelope protein precursor no longer enters the secretory pathway. If the cDNA fragment fused to the mutated precursor contains a signal peptide, membrane localization of the envelope proteins is restored. If this happens, the encapsidated viral genome is released within an infectious viral particle in the culture medium. On the other hand, if the cDNA fragment inserted in the viral genome does not encode a signal peptide, then envelope proteins do not enter the secretory pathway and no viral particle is produced. Thus, the selection step is done by the cell itself. The recovery of infectious viral particles in the culture medium is a rapid and efficient way to isolate nucleic acids encoding a specific function (e.g. secretory signal) from a complex library. We call this method the expression-based virus selection system (EB-VSS).
MATERIALS AND METHODS
Plasmid construction
The p62mut Sindbis virus genome was constructed as follows. The mouse growth hormone cDNA (mGH, GenBank accession no. X02891) was amplified by RT–PCR using total RNA from mouse pituitary and forward primer 5′-agcgaattcgtcctgtggacagatcactgc-3′ and reverse primer 5′-gctctcgaggaaggcacagctgctttccac-3′. The mGH product was cloned in pcDNA1.1 (Invitrogen, Carlsbad, CA). Part of the Sindbis virus p62-6K-E1 coding sequence was amplified from plasmid DH-BB (Invitrogen) using primers 5′-cttctcgagcagtttaaacgtgagcttccc-3′ and 5′-acgtctagatcatcttcgtgtgctagtcag-3′. The resulting product, lacking the first 13 amino acids of p62, was cloned downstream and in-frame with mGH in pcDNA-mGH. A double-stranded oligonucleotide (5′-tcgagcagatctgcagcaccactggtcacggcaatgtgtcggagcgg-3′/5′-ccgctccgacacattgccgtgaccagtggtgctgcagatctgc-3′) was inserted between the mGH and p62Δ13 coding sequences to reconstitute p62mut. The capsid protein coding sequence was amplified from DH-BB using forward (5′-gtgtccaagccatcagaggggaaataaagcatctctacggtggtcctaaatagtcagcatagt-3′) and reverse (5′-ccagagctcatgcggaccactcttctgt-3′) primers. The forward primer contains nucleotides –46 to +14 of the Sindbis virus subgenomic mRNA promoter. The reverse primer contains a termination codon. The PCR product was cloned in a blunted XbaI site of pcDNA-mGH-p62mut-6K-E1. The entire mGH-p62mut-6K-E1-C cassette was then excised and subcloned at the unique PmlI site of a Sindbis virus replicon inserted in pcDNA1.1. The Sindbis virus replicon was excised from the pSinRep5 plasmid (Invitrogen). A unique SwaI site was introduced at the 3′-end of the viral sequence by PCR mutagenesis. Finally, oligonucleotide adapters inserting unique restriction sites (NotI and BamHI) were cloned in place of mGH to allow subcloning of cDNA fragments upstream of p62mut. A fragment of the mouse keratin-associated protein (mKAP, GenBank accession no. AF031485) was amplified by RT–PCR using total RNA from mouse embryonic E16.5 skin and forward primer 5′-gcggccgcgagaattcgtctcctctcaacatggtctac-3′ and reverse primer 5′-caggatcctgaggagggcagg-3′. The mKAP product was then subcloned in place of mGH in the above described construct. The green fluorescent protein (GFP) coding sequence (mutant F64L, S65C and I168T) was obtained from plasmid pQBI25fc3 (Quantum Biotechnologies, Montreal, Canada). A minimal subgenomic mRNA promoter (–46/+14) was added at the 5′-end and the resulting transcription unit was inserted between the structural proteins coding sequence and the 3′-untranslated region of a Sindbis virus genome.
cDNA library construction
Libraries of cDNA fragments were constructed according to the oligo-capping method (7). Briefly, poly(A)+ RNA (2–5 µg in 100 µl) was treated with 1.2 U of bacterial alkaline phosphatase (Takara) for 1 h at 37°C. After purification, the RNA preparation was treated with 10 U of tobacco acid pyrophosphatase (Epicentre, Ontario, Canada) for 1 h at 37°C and then ligated overnight at 12°C to an oligoribonucleotide (5′-gaccuaagcaucgagugcggccgcuacgaa-3′) using 100 U of T4 RNA ligase (New England Biolabs) in a volume of 100 µl. First strand cDNA synthesis was performed at 42°C using 600 U of Superscript II (Invitrogen) and a N9 linker-primer (5′-gagagagagagagcgactcggatccannnnnnnnnc-3′). PCR (20– 30 cycles) was performed using Taq DNA polymerase (Amersham Pharmacia), reverse primer (5′-gagagcgactcggatcca-3′) and forward primer (5′-ctaagcatcgagtgcggc-3′). After digestion with NotI and BamHI, PCR products were size selected (300–800 bp) by agarose gel electrophoresis and cloned in an equimolar mix of three p62mut Sindbis virus replicons. Each of the three replicons contains p62mut in a different frame relative to the cloning sites.
In vitro transcription
Plasmids linearized by SwaI were used as template for transcription of capped RNA using the RiboMax SP6 kit (Promega, Madison, WI). Synthetic RNA was purified using RNeasy columns (Qiagen).
Cell culture, transfection and infection
BHK-21 cells (Invitrogen) were grown in α-MEM (Life Technologies) supplemented with 5% fetal bovine serum. Plasmid DNA (1 µg) was transfected using the FuGENE-6 reagent (Roche Molecular, Laval, Quebec, Canada) according to the manufacturer’s instructions. RNA (8–10 µg) was transfected by electroporating 10 × 106 BHK-21 cells using a Bio-Rad GenePulser II set at 1.25 kV, 25 µF and maximal resistance [modified from Liljeström et al. (8)]. Medium was changed after 16 h and left to accumulate viral particles for 4 h. The medium was filtered through a 0.45 µm membrane and the viral titer was determined. BHK-21 cells (10 × 106) were infected at a maximal m.o.i. of 0.1 for 2 h and the medium was changed. Infected cells were processed after 20 h.
RNA extraction and RT–PCR
Total cellular RNA was purified using the RNeasy kit (Qiagen). RT–PCR was performed using the Titan one tube RT–PCR system (Roche Molecular). Products of the RT–PCR reaction were cloned in pBluescriptKS-II (Stratagene) and inserts from isolated colonies were sequenced. Sequences were compared to GenBank entries using Blastn. The presence of a signal peptide was determined using the SignalP server V2.0.1b (9).
Production of recombinant proteins and polyclonal antibodies
The full-length C protein coding sequence was amplified from DH-BB (Invitrogen), cloned in the EcoRV site of pBLUESCRIPT KS™ (Stratagene, La Jolla, CA). A HindIII–BamHI fragment from the resulting plasmid was cloned into prokaryotic expression plasmid pQE30 (Qiagen, Mississauga, Ontario, Canada) in-frame with an N-terminal hexahistidine tag. The E1 protein coding sequence was amplified from DH-BB. The resulting 1320 bp PCR product was cloned into pQE30 and the resulting plasmid was digested with StuI and HindIII, blunted and recircularized. The resulting construct contains amino acids 1–340 of E1 fused to an N-terminal hexahistidine tag. Fusion proteins were produced in Escherichia coli strain SG13009(pREP4) and purified under denaturing conditions according to the manufacturer’s instructions (QiaExpressionist™ kit; Qiagen). The proteins were further purified on denaturing polyacrylamide gels. The gel-extracted proteins were precipitated with trichloroacetic acid (10% v/v), resuspended in 8 M urea and dialyzed at 4°C against 4 l of TBS. Aliquots of 200 µg of recombinant protein mixed with complete Freund’s adjuvant (VWR Canlab, Montreal, Quebec, Canada) were injected s.c. into New Zealand White rabbits on day 1. On days 15 and 28, another 100 µg of recombinant protein mixed with incomplete Freund’s adjuvant was similarly injected. Rabbits were bled 7 days after the last injection.
Detection of E1 protein
Cell surface proteins were biotinylated using sulfo-NHS-biotin (0.5 mg/ml) as recommended by the manufacturer (Pierce, Rockford, IL). Membrane proteins were solubilized in lysis buffer (1% IGEPAL-630, 50 mM Tris–HCl pH 8, 150 mM NaCl, 2 mM EDTA) and separated by SDS–PAGE. Western analysis was performed using a 1/500 dilution (v/v) (final concentration 1 µg/ml) of affinity-purified anti-E1. Detection was performed with the ECL reagent (Amersham Pharmacia Biotech, Baie d’Urfé, Quebec, Canada) after incubation with a 1/10 000 dilution (v/v) (final concentration 80 ng/ml) of goat anti-rabbit IgG coupled to horseradish peroxidase (Sigma, St Louis, MO).
Immunofluorescence
Cells grown on coverslips were fixed with 2% paraformaldehyde (w/v) and sequentially incubated with a 1/50 dilution of the anti-C antiserum and a 1/150 dilution of goat anti-rabbit IgG coupled to tetramethyl rhodamine isothiocyanate (Sigma).
Determination of viral titer
BHK-21 cells (10 × 106) were electroporated with 10 µg of replicon RNA, seeded and left for 16 h. After rinsing the cells, fresh medium was added and collected after 3 h. The medium was filtered through a 0.45 µm membrane and serial dilutions were used to infect fresh BHK-21 cells for 4 h. Cells were then overlaid with Seaplaque agarose (0.9% w/v) and viral plaques were counted 48 h later.
RESULTS
Our novel functional genomics tool uses engineered viral genomes to express a library of nucleic acid fragments in mammalian cells and selectively retrieve those encoding a specific function. Specific cDNA or cDNA fragments can be retrieved from complex libraries by virtue of being able to complement a defective viral function. The principle of our methodology is genetic complementation. A similar approach was developed by Sleat et al. to show that foreign gene transcripts could be recovered as virus-like particles in virus-infected transgenic tobacco (10). The nature of the viral defect dictates the function being screened for. In the present report, we describe the application of this technology to identify cDNAs encoding secreted and membrane-bound proteins. cDNA fragments are screened for the presence of a signal peptide able to restore membrane localization of viral envelope proteins.
Design of a signal trap using expression-based virus selection system
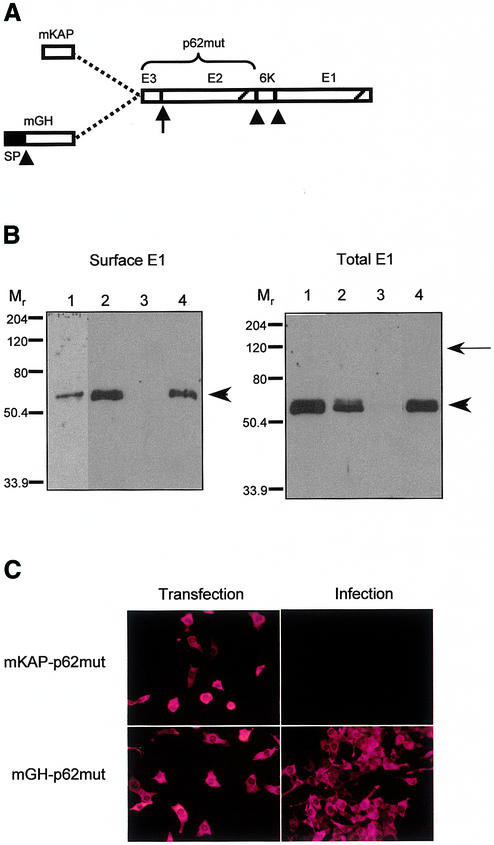
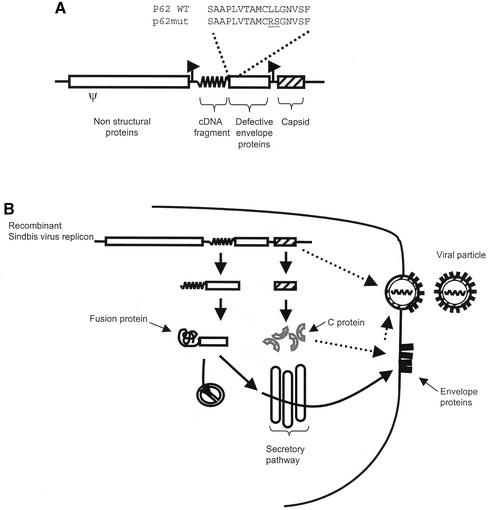
We have chosen to use Sindbis virus replicons as our expression/selection vector. Sindbis virus is a positive-strand RNA alphavirus with a broad host range (11). It is well characterized and its relatively small genome (11.7 kb) can be easily manipulated by genetic engineering. Copies of the genome synthesized in vitro are infectious and can express heterologous nucleic acids in mammalian cells (12). The genomic RNA encodes non-structural proteins responsible for its replication and for the synthesis of a subgenomic mRNA from an internal RNA promoter (13). The subgenomic mRNA encodes the structural proteins, namely C, p62, 6K and E1, which are synthesized as a polyprotein precursor. The C protein co-translationally cleaves itself from this precursor and coats the genomic RNA to form the nucleocapsid. Cleavage of the C protein uncovers the N-terminal signal peptide of the p62 protein, which directs the remaining portion of the precursor to the secretory pathway (14). Action of the signal peptidase generates the p62, 6K and E1 moieties. p62 and E1 heterodimerize in the endoplasmic reticulum (15). The N-terminal region of p62 is thought to play a transient role in this assembly (16). The signal peptide of p62 is not cleaved; the protein is processed along the secretory pathway to generate E2 and a 64-residue N-terminal fragment (designated E3). E3 is not retained in the mature Sindbis virus particle (16). E2 and E1 constitute the viral envelope. Therefore, production of Sindbis virus particles is dependent upon the entry of p62 into the secretory pathway. We reasoned that it could be possible to develop a signal trap by fusing heterologous nucleic acids to the 5′ end of p62. However, we found that the signal peptide of p62 could act internally when fused to short N-terminal extensions (Fig. 2B, lane 1; see below). To prevent entry of p62 into the secretory pathway, we therefore substituted two residues within the hydrophobic core of its signal peptide (L11R and L12S, designated p62mut). These mutations were introduced in an attempt to block the signal peptide function of the N-terminal region of p62 without affecting its role in the heterodimerization of p62 and E1. Before cloning cDNA fragments upstream of p62mut, it was necessary to engineer the Sindbis virus replicon such that the synthesis of the C protein be uncoupled from the synthesis of the envelope proteins. Indeed, the C protein coding sequence is found upstream and in-frame with p62 in the wild-type alphaviral genome. It has been reported that Sindbis virus replicons can harbor more than one subgenomic RNA promoter (17). We took advantage of this property and placed the C protein coding sequence under the control of an additional subgenomic mRNA promoter. Finally, cloning sites were introduced upstream of the p62mut coding sequence to allow insertion of cDNA fragments. The Sindbis virus replicon engineered to retrieve cDNA fragments encoding secreted or membrane-bound proteins is schematized in Figure 1A.
Figure 2.
(A) Schematic representation of the mutated envelope polyprotein precursor fused to a cDNA fragment either containing a signal peptide (mGH) or not (mKAP). Arrowheads indicate cleavage by signal peptidase; arrow indicates site of processing of p62mut in the trans-Golgi network; diagonals represent transmembrane domains. (B) Cell surface (left) and total (right) E1 revealed by western analysis 24 h after transfection in BHK-21 cells of CMV-based plasmids expressing p62WT fused either to mKAP (lane 1) or mGH (lane 2) and p62mut fused either to mKAP (lane 3) or mGH (lane 4). Arrowheads indicate the position of the E1 protein; the arrow points to the expected position of the unprocessed mKAP-p62mut putative polyprotein. (C) Anti-C immunostaining performed on cells 12 h after transfection with p62mut Sindbis virus replicon containing the mKAP fragment (top left) or the mGH fragment (bottom left). Right panels show anti-C immunostaining of cells infected with culture medium from the corresponding transfected cells.
Figure 1.
(A) Signal sequence trap expression and selection vector. The insert (wiggled line) is fused to p62mut. Amino acid substitutions (underlined) within the signal peptide of p62 are indicated. The mRNA encoding the C protein (hatched box) is expressed from a distinct subgenomic mRNA promoter. Ψ, putative encapsidation signal; arrows, subgenomic mRNA promoters. (B) Once transfected into a cell, the recombinant Sindbis virus replicon expresses two mRNAs. Translation of these mRNAs produces the C protein and a chimeric protein consisting of an N-terminal portion encoded by the cDNA fragment fused to the p62mut envelope protein precursor. Depending on the presence of a signal peptide, the chimeric protein remains cytosolic or co-translationally enters the secretory pathway, where the envelope proteins are normally synthesized and processed and ultimately reach the plasma membrane. There, assembly of the envelope proteins with the nucleocapsid leads to budding of a viral particle.
Figure 1B depicts the selection process occuring in cells transfected with such replicons. Upon entry into the cell, the replicon is translated into non-structural proteins which eventually synthesize two distinct mRNAs from the corresponding subgenomic mRNA promoter. One of these encodes the C protein which encapsidates the replicon. The other subgenomic mRNA is translated into a fusion protein consisting of an N-terminal region encoded by the heterologous cDNA fragment followed by p62mut-6K-E1. If the cDNA fragment does not encode a signal peptide, then an aberrant and non-functional fusion protein is produced. On the other hand, if the cDNA fragment encodes a signal peptide, then the fusion polyprotein enters the secretory pathway and is processed into E2/E1 envelope proteins that ultimately reach the cell surface. Only transfected cells displaying envelope proteins at the cell surface can release infectious viral particles into the culture medium. As can be seen, the engineered Sindbis virus replicon serves both as an expression vector and a selection tool.
To test whether the biosynthesis of p62mut-6K-E1 was restored when fused to a signal peptide-containing moiety, the coding sequence of mGH (675 bp encoding 216 residues) was cloned upstream of p62mut. A fragment containing an initiator methionine but lacking a signal peptide (residues 1–24 of a mouse keratin-associated protein, mKAP) was used as a negative control. These constructs, as well as the wild-type counterparts (mGH-p62WT and mKAP-p62WT), were initially tested in the context of CMV-based DNA plasmids. The resulting fusion proteins (heterologous fragment + p62mut) are schematized in Figure 2A. After transfection in BHK-21 fibroblasts, cell surface biotinylation and western analysis were performed in order to detect the presence of E1 at the cell surface. Localization of E1 at the cell surface is indicative of normal biosynthesis of envelope proteins, i.e. entry of p62 and E1 into the secretory pathway and transport to the plasma membrane as heterodimers. Fusion of mGH to p62mut was able to restore entry of p62mut in the secretory pathway and it did not impair the biosynthesis and heterodimerization of envelope proteins, as evidenced by the cell surface localization of E1 (Fig. 2B, lane 4, left panel). As expected, the mKAP fragment failed to restore localization of E1 at the cell surface (Fig. 2B, lane 3, left panel). When fused to p62WT, both inserts led to E1 at the cell surface (Fig. 2B, lanes 1 and 2, left panel). Taken together, these results indicate that: (i) the signal peptide of p62 can function internally when fused to a short N-terminal extension; (ii) a protein that is fused at the N-terminus of p62 (WT or mut) does not prevent biosynthesis/processing of the envelope proteins. Interestingly, total cellular E1 was barely detectable when expressed from the mKAP-p62mut construct (Fig. 2B, lane 3, right panel). Northern analysis confirmed that similar levels of mKAP-p62mut and mGH-p62mut mRNAs were expressed (not shown). It is unlikely that the mKAP fragment affects replication or translatability of the recombinant viral genome since it has no effect in the context of p62WT (Fig. 2B, lane 1, right panel). The lack of a signal peptide in p62mut should in theory result in the production of a larger 110 kDa cytosolic polyprotein (Fig. 2B, arrow). However, this putative protein would contain multiple highly hydrophobic (transmembrane) domains (see Fig. 2A). These would trigger misfolding and/or aggregation of the protein which would be rapidly degraded. This could explain the absence of any cellular E1 signal after transfection of the mKAP-p62mut construct.
In order to assess the specificity of our signal trap assay, engineered Sindbis virus replicons containing either a mKAP or mGH cDNA fragment were electroporated into BHK-21 fibroblasts. Twelve hours post-transfection, cells were fixed and the medium was collected and used to infect fresh monolayers of BHK cells. Infected cells were fixed 20 h later. Anti-C protein immunofluorescence was performed on transfected and infected cells (Fig. 2C). The C protein serves as a marker to determine transfection efficiency and to detect the presence of viral plaques. Only the replicon containing mGH led to production of infectious viral particles (Fig. 2C, bottom right panel), whereas mKAP did not (Fig. 2C, top right panel). Taken together, these results indicate that a cDNA encoding a functional signal peptide can complement the defect of the p62mut-containing Sindbis virus replicon. Viral production was quantified as follows. Cells were electroporated with replicon RNA, seeded and incubated for 13 h. At this time, medium was changed and left on the cells for 3 h. This viral stock was titered by counting plaque-forming units (p.f.u.) after agarose overlay. In this assay, mGH-containing replicons gave rise to 1.2 × 103 p.f.u. Under similar conditions, a replicon containing the wild-type structural proteins precursor and a second transcription unit expressing GFP gave rise to 1.4 × 105 p.f.u. The difference in the amount of plaque-forming units released from cells transfected with mGH-p62mut or wild-type replicons is mainly due to the lowered expression of envelope proteins. Indeed, a powerful translation enhancer is present in the 5′ region of the C protein coding sequence in the wild-type structural proteins mRNA (18). In the mGH-p62mut replicon, this enhancer has no effect on the translation of the envelope proteins precursor mRNA because the C protein is expressed from a distinct subgenomic mRNA promoter (see Fig. 1).
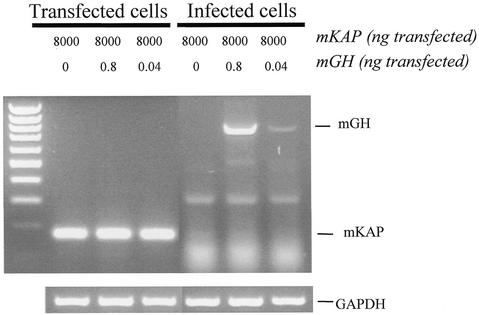
We next sought to determine the sensitivity of our assay. In order to do so, a fixed amount of mKAP-p62mut replicon (8 µg) was spiked with decreasing amounts of mGH-p62mut replicon (0.8 and 0.04 ng). These dilutions (1/10 000 and 1/200 000 w/w) were electroporated into BHK-21 fibroblasts. Medium was changed 20 h post-transfection and left on the cells for 3 h to accumulate viral particles. This medium was used to infect a fresh monolayer. RT–PCR performed with primers flanking the insert revealed only the presence of the mKAP insert in total RNA extracted from transfected cells (Fig. 3, lanes 2–4). The mGH insert was not detected in RNA from transfected cells owing to its very low abundance and to the fact that it is competed out by the mKAP insert. Indeed, the latter is expressed in much greater amount from a vast excess of transfected replicons. However, RT–PCR performed on total RNA from infected cells revealed only the mGH fragment, which was still readily visible by ethidium bromide staining even when initially transfected at a 1/200 000 dilution (Fig. 3, lane 7). These results are consistent with the immunofluorescence data presented in Figure 2C and clearly demonstrate that a fragment encoding a signal peptide (mGH) can be specifically retrieved when co-transfected with up to 200 000 times more of a fragment not encoding a signal peptide (mKAP). We have found that the length of the fusion to p62mut can influence the efficiency of viral particle production. Indeed, fusion of a truncated form of mGH encoding only its signal peptide (29 residues fused to p62mut) resulted in a 5-fold increase in release of viral particles when compared to its full-length counterpart (216 residues fused to p62mut). This indicates that the sensitivity determined using full-length mGH (1/200 000) is a conservative estimate.
Figure 3.
Sensitivity of EB-VSS signal sequence trap. Results of RT–PCR on RNA from cells transfected (lanes 2–4) with different ratios of p62mut Sindbis virus replicons containing either a positive insert (mGH) or a negative insert (mKAP) or from cells infected (lanes 5–7) with the corresponding cell culture medium. Amplification of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used as a control (bottom panel). The left lane shows migration of a 100 bp marker.
Construction and screening of libraries of cDNA fragments
Signal peptides are located at the N-terminus of proteins destined for the secretory pathway and are therefore encoded by the 5′ region of cDNAs. Analysis of sequences of 68 known secreted or membrane-bound proteins (taken from GenBank) showed that the signal peptide is found within the first 400 bp of the corresponding mRNA in all cases. As reported by others using signal trap assays, screening of cDNA fragments randomly generated yields a high rate of false positives (20–50%) due to fragments fortuitously encoding a methionine followed by a stretch of hydrophobic amino acids, which can act as weak signal peptides (19). Furthermore, because of the limitations of reverse transcriptase, the 5′ region of cDNAs are under-represented in libraries of randomly generated cDNA fragments. We therefore adapted the ‘oligo-capping’ method to construct high quality libraries enriched in 5′ fragments of cDNAs (7). The oligo-capping technique relies on the property of tobacco acid pyrophosphatase to specifically convert the CAP structure of full-length mRNA to a ligatable 5′ phosphate moiety. The addition of an oligoribonucleotide primer at the 5′-ends of mRNA and the subsequent synthesis of first strand cDNA using a linker-random primer allows the amplification of 5′ regions of cDNAs. Amplicons are size selected (300–800 bp) and directionally cloned in an equimolar mix of three engineered replicons upstream of p62mut. To maximize the probability of in-frame fusion, each of the three replicons contains p62mut in a different frame relative to the cloning sites. Table 1 outlines the characteristics of sample libraries. Typical libraries contain 3 × 105 to 8 × 105 clones. Random sequencing revealed that more than 80% of inserts with homology to known sequences correspond to the 5′ region of the cDNA.
Table 1. Characterization of sample cDNA fragment libraries.
| Tissue sourcea | mRNA (µg) | Clones (n) | Percent 5′ regionb (n) |
|---|---|---|---|
| Proximal limb buds (E12.5) | 3.0 | 3.8 × 105 | 84 |
| Ribs (E16.5) | 4.6 | 4.8 × 105 | 91 |
| SaOS-2 cell line | 4.0 | 4.0 × 105 | 89 |
| Limb buds (E14.5) | 3.0 | 5.9 × 105 | 64 |
| UMR-106 cell line | 4.3 | 7.3 × 105 | 85 |
aTissues are dissected from mouse embryos at the indicated stage.
bPercentage of inserts with homology to known sequences corresponding to the 5′ region of the cDNA, determined from random sequencing of 40–48 clones.
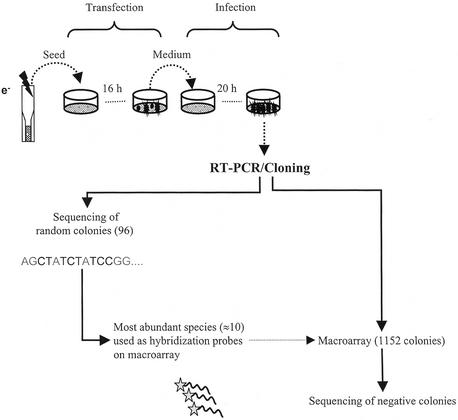
RNA copies of recombinant replicons are synthesized in vitro and electroporated into BHK-21 fibroblasts. Typically, 50 million cells are transfected with an efficiency of at least 10%. A 10-fold coverage of the library (5 million assays) is thus achieved in a single experiment. The medium of transfected cells is changed 16 h after seeding and left on the cells for 4 h to accumulate viral particles. This medium is then used to infect a fresh monolayer of BHK cells. Total RNA is extracted 20 h later. cDNA fragments found in the infected cells are amplified using primers flanking the site of insertion in the engineered replicon. Amplicons are shotgun cloned in a bacterial plasmid. Hence, the population of inserts contained within viral particles is transformed into a population of bacterial clones. Inserts from 96 randomly chosen colonies are sequenced to evaluate the diversity of retrieved sequences and to identify the most abundant species. These are radiolabeled and hybridized to a 1152-colony macroarray of randomly chosen bacterial colonies. Inserts from negative colonies (typically representing 30–35% of the macroarray) are sequenced. Figure 4 schematizes the screening procedure.
Figure 4.
Experimental protocol for library screening and analysis of contents of viral particles. A library of Sindbis virus replicons is electroporated into BHK-21 fibroblasts. Culture medium is changed after 16 h and left on the cells for 4 h to accumulate viral particles. This medium is used to infect cells. RNA is extracted 20 h post-infection. RT–PCR is performed using primers flanking the site of insertion of cDNA fragments. Amplicons are shotgun cloned in a bacterial plasmid. A macroarray of 1152 bacterial colonies is made. Inserts from 96 randomly picked colonies are sequenced. The most abundant inserts (between 5 and 10 clones) are labeled and hybridized to the macroarray. Inserts from negative colonies are sequenced.
Table 2 summarizes the results obtained from screening various libraries of cDNA fragments. These libraries were constructed from RNA extracted from the developing skeleton (five libraries), adult bone (one library), adult bone marrow (one library) and osteoblastic cell lines (two libraries). A total of 46 million assays (transfected cells) were performed. 9463 bacterial colonies were analyzed either by sequencing or hybridization. 2546 inserts were sequenced, 97.1% of which encoded a signal peptide as determined using the SignalP bioinformatics software (9). These 2473 inserts represented 419 distinct cDNA species. Seventy-three inserts (2.9%) did not contain a bona fide signal peptide. These were derived from sequences fortuitously encoding a stretch of hydrophobic amino acids preceded by an initiator ATG. Of the 419 cDNA species retrieved, 325 (77%) were identical or closely related to known proteins. Included were secreted proteins and type I and type II integral membrane proteins, as well as multipass and GPI-anchored proteins. Table 3 lists some of the annotated cDNA species that were retrieved from the libraries. It should be noted that EB-VSS can retrieve cDNA fragments derived from mRNA expressed at very low levels in the starting material (e.g. leptin receptor and endothelin receptor type B). Seventy-one cDNA species corresponded to annotated proteins of unknown function and 23 species were either entirely novel proteins or not predicted to contain a secretory signal from available sequence data. Table 4 gives some examples of the latter. Taken together, these results indicate that EB-VSS efficiently filters complex cDNA libraries to retrieve only those encoding secreted or membrane-bound proteins.
Table 2. Summary of screening results.
| Library | Assaysa | Bacterial colonies analyzed | Inserts sequenced | Inserts encoding SP | Unique cDNAs annotated | Unique cDNAs unannotatedb |
|---|---|---|---|---|---|---|
| 1 | 3 × 106 | 1252 | 300 | 293 | 55 | 17 |
| 2 | 6 × 106 | 1252 | 248 | 247 | 81 | 21 |
| 3 | 3 × 106 | 75 | 75 | 74 | 24 | 2 |
| 4 | 5 × 106 | 1252 | 385 | 382 | 106 | 17 |
| 5 | 8.5 × 106 | 1252 | 435 | 418 | 112 | 19 |
| 6 | 5 × 106 | 1252 | 220 | 219 | 64 | 5 |
| 7 | 3 × 106 | 624 | 112 | 107 | 22 | 0 |
| 8 | 6.2 × 106 | 1252 | 312 | 290 | 65 | 24 |
| 9 | 6.3 × 106 | 1252 | 459 | 443 | 84 | 20 |
| Total | 46 × 106 | 9463 | 2546 | 2473 | ||
| Non-redundant | 325 | 94 |
aCorresponds to the estimated number of transfected cells, as determined by immunofluorescence.
bEncodes novel protein or protein of unknown function.
Table 3. Examples of retrieved cDNAs encoding secreted or membrane-bound proteins.
| Gene product | GenBank accession no. | Retrieved fragmenta | Typeb | Abundancec |
|---|---|---|---|---|
| Bone sialoprotein | NM008318 | –71/+495 | Sec | High |
| Calumenin | NM007594 | –63/+189 | Sec | High |
| Cathepsin K | AK003425 | –52/+122 | Sec | Medium |
| CD45, Tyr phosphatase receptor | XM016748 | –81/+182 | I | Medium |
| Collagen type Iα2 | NM007743 | –80/+535 | Sec | High |
| Dickkopf-1 | AF030433 | –22/+442 | Sec | Medium |
| Endothelin receptor type B | NM007904 | –203/+166 | Mp | Low |
| GPCR5C orphan receptor | AK014308 | –33/+106 | Mp | Low* |
| Hedgehog interacting protein | NM020259 | –182/+130 | I | Low |
| Hypoxia-induced gene 2 | AK009377 | –191/+185 | II | Medium* |
| Leptin receptor | NM010704 | –131/+69 | I | Low |
| Lysosomal proton pump Atp6s | NM031785 | –2/+241 | I | High |
| Nicastrin | AF240469 | –18/+201 | I | Medium |
| Osteocalcin | X04142 | –47/+269 | Sec | High* |
| Osteopontin | J04806 | –69/+303 | Sec | High |
| Resistin-like α | AF323082 | –76/+123 | Sec | Medium |
| Sclerostin | AK017295 | –39/+158 | Sec | Low* |
| Signal peptidase SPC22 | AK007432 | –58/+191 | II | High |
| TMS-1 | AF181684 | –34/+180 | Mp | Medium |
a+1 corresponds to initiator ATG.
bSec, secreted; I, type I integral membrane protein; II, type II integral membrane protein; Mp, multipass membrane protein.
cRelative abundance of corresponding mRNA in starting material, approximated from published data or according to actual northern analysis (*).
Table 4. Examples of misannotated or unannotated cDNAs retrieved using EB-VSS.
| Clone name | GenBank match | Region of homology | Putative sequencea | SignalP scoreb (%) | Comments |
|---|---|---|---|---|---|
| Saos-cap1b-c1-neg57 | AF161372 | 945–724 | MASLLCCGPKLAACGIVLSAWGVIMLIMLGIFFNVHSAVLIEDVPFTEKDFENG | 95.3 (anchor) | Reading frame in opposite orientation from the one predicted |
| Ca155-cap1-c2-neg052 | AK002812 | 2–481 | MPHAALSSLVLLSLATAIVADCPSSTWVQF… | 99.9 | Sequencing errors, wrong initiator ATG used to predict open reading frame |
| Lmb145-cap1-c-410 | BB590753, AY100450c | – | MCAPGYHRFWFHWGLLLLLLLEAPLRGLALPPIRYSHAGICPN | 99.9 | Small EST (153 bp) does not contain signal peptide |
| Ca155-cap1-c2-neg228 | AY100451c | – | MGALQNVSRFFFLVTFMFGDAIFTLPSSTIRSGFAPYATAMAINLTQQLASP | 88.2 | No match in databases |
| Lmb145-cap1-c-243 | AY100452c | – | MERPQSSIWVFMLLLFMVLLQSPAWHVAAQRCPQTCVCDNSRRHVTCRHQNLTEVPNTIPELTQRLDLQGNILKVLPAAAFQ | 99.9 | Novel protein |
aDeduced from the retrieved fragment sequence. The predicted signal peptide or anchor is underlined.
bDetermined according to Nielsen et al. (9).
cFrom this work.
DISCUSSION
The rationale behind our novel expression screening system is to link the expression and the retrieval of a cDNA encoding a specific function. This is achieved by expressing cDNAs from a recombinant alphaviral genome able to package itself only if the cDNA it expresses can complement a specific defect in viral particle formation. In the application reported here, we have shown that a defect in viral particle formation due to mutations in the signal peptide of the Sindbis virus p62 envelope protein can be rescued by fusion to a cDNA encoding a signal peptide. This adaptation of the ‘signal trap’ strategy presents many advantages. Since there is no need to select cells expressing the target cDNA fragments, our method is high throughput. Indeed, packaging of target cDNAs within viral particles allows retrieval simply by collecting the cell culture medium, typically 24 h after transfection of a library. The method is also very sensitive because the viral particles are infectious and the engineered viral genome is replication-competent. It is estimated that replication of the Sindbis virus genome inside an infected cell gives rise to 130 000–160 000 copies of the replicon within 8 h after infection (20). This property allowed us to easily retrieve as little as 40 pg of positive Sindbis virus replicon transfected into BHK-21 fibroblasts along with 200 000-fold more negative Sindbis virus replicon. Importantly, no viral revertant was ever found from cells transfected with a Sindbis virus replicon in which a cDNA fragment not encoding a signal peptide was fused to p62mut. This is presumably due to the fact that the viral replicon has been extensively rearranged, i.e. mutations in the p62 signal peptide, insertion of heterologous cDNA fragments and displacement of the C protein coding sequence. To our knowledge, no other reporter system matches the level of signal amplification achieved through viral infection/replication. The limit of detection of signal traps using enzymatic or fluorescent reporters in mammalian cells has been reported to range between 1/200 and 1/5000 (5,21,22).
In order to identify putative bone anabolic agents, we have screened various libraries of cDNA fragments derived from adult and developing bone as well as from osteoblastic cell models. A total of 2546 sequences yielded 419 unique cDNAs encoding secreted or membrane-bound proteins. The retrieval of several fragments encoding uncleaved signal anchor found in type II integral membrane proteins (see Table 3 for examples) suggests that a variety of fusion partners are tolerated at the N-terminus of p62mut without deleteriously affecting processing and transport of envelope proteins. It should be noted that viral particles produced in our assay contain wild-type E2 since the chimeric p62mut proteins are cleaved at the E3/E2 junction as part of the normal maturation of Sindbis virus envelope proteins. Our libraries contain fragments ranging from 300 to 800 bp in length. This fragment size should be well tolerated by the viral machinery since it has been reported that a recombinant Sindbis virus genome containing 1.5 kb of heterologous sequence was efficiently packaged and released within infectious viral particles (17).
Analysis of screening results indicated that less than 3% of inserts did not encode true signal peptides. This is to be compared to a rate of false positives of 20–50% obtained using other techniques (5,21,22). The improved performance of our system can be explained by the fact that (i) more than 80% of the cDNA fragments present in the libraries correspond to the 5′ region of mRNA, thereby eliminating artefactual signal peptides arising from random fragments encoding a methionine followed by a stretch of hydrophobic residues; (ii) no cell selection step is involved; (iii) release of infectious viral particles is stringently dependent on the fusion of a functional signal peptide to p62mut as we have found that the envelope proteins do not even accumulate in its absence (see Fig. 2).
A potential limitation of our methodology is that it relies on a number of amplification steps that may introduce artefactual biases. This is especially true during library construction. Clearly, the use of PCR to construct libraries of cDNA fragments shifts the representation of clones towards more abundant mRNAs. However, this bias could be reduced by normalizing the mRNA population before screening. We also found some variation (2–5-fold) in the efficiency of viral particle formation from different Sindbis virus replicons, presumably due to the varying strength of individual signal peptides encoded by heterologous cDNA fragments. Despite the previous observations, we have been able to retrieve cDNA fragments derived from low abundance mRNAs, e.g. encoding seven transmembrane domain receptors (see Table 3 for examples).
Because of their biological importance and therapeutic potential, genomic data are being extensively screened for secreted and membrane-bound proteins (23). Large-scale EST sequencing coupled to in silico prediction of signal peptides has proven to be a powerful tool in this task (24). Nonetheless, our functional approach has revealed a number of cDNAs that were not annotated as containing a signal peptide in the databases, either because the 5′ region was missing or due to the presence of sequences disrupting the open reading frame (small introns and sequencing artefacts). In fact, we succeeded in identifying novel secreted and membrane-bound proteins at a relatively high rate (24% of non-annotated cDNA that were retrieved). Furthermore, 2546 sequencing reactions yielded 419 unique signal peptide-containing cDNAs, an average of only 6.1 sequences per cDNA. These findings illustrate the power of filtering genetic information in mammalian cells.
Recombinant viruses have been used for some time to introduce nucleic acids into mammalian cells for expression screening purposes, i.e. to serve as a general gene ‘delivery’ vehicle (25,26). Our work has shown that viruses can also be engineered to selectively get nucleic acids out of cells, i.e. to serve as a specific gene ‘recovery’ vehicle. We engineered Sindbis virus replicons to specifically recover cDNA fragments encoding secretory signals. Others had previously shown that recombinant viruses can be used to screen libraries of oligonucleotides for those able to substitute for the Sindbis virus subgenomic mRNA promoter (27) or encoding putative protease cleavage sites. For the latter, Buchholz et al. fused EGF to the envelope glycoprotein of a retrovirus and randomized the linker region to screen for sites that can be cleaved by endogenous cellular proteases (28). In a more specific screening assay, Pacini et al. engineered Sindbis virus replicons to retrieve sites cleaved by the hepatitis C virus serine protease (NS3-4Ap) (29,30). With the recent flood of genomics data, it is now clear that new tools are needed to rapidly and efficiently decipher gene function in living mammalian cells. We believe the use of engineered viruses to recover nucleic acids encoding specific functions can greatly increase the throughput of expression screening methodologies since it bypasses the limiting step of having to select individual mammalian cells.
Acknowledgments
ACKNOWLEDGEMENT
We thank Jacques Drouin (Institut de Recherches Cliniques de Montréal) for helpful advice.
DDBJ/EMBL/GenBank accession nos AY100450–AY100452
REFERENCES
- 1.Walter P., Gilmore,R. and Blobel,G. (1984) Protein translocation across the endoplasmic reticulum. Cell, 38, 5–8. [DOI] [PubMed] [Google Scholar]
- 2.Tashiro K., Tada,H., Heilker,R., Shirozu,M., Nakano,T. and Honjo,T. (1993) Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science, 261, 600–603. [DOI] [PubMed] [Google Scholar]
- 3.Klein R.D., Gu,Q., Goddard,A. and Rosenthal,A. (1996) Selection for genes encoding secreted proteins and receptors. Proc. Natl Acad. Sci. USA, 93, 7108–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojima T. and Kitamura,T. (1999) A signal sequence trap based on a constitutively active cytokine receptor. Nat. Biotechnol., 17, 487–490. [DOI] [PubMed] [Google Scholar]
- 5.Chen H. and Leder,P. (1999) A new signal sequence trap using alkaline phosphatase as a reporter. Nucleic Acids Res., 27, 1219–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seed B. (1995) Developments in expression cloning. Curr. Opin. Biotechnol., 6, 567–573. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki Y., Yoshitomo-Nakagawa,K., Maruyama,K., Suyama,A. and Sugano,S. (1997) Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene, 200, 149–156. [DOI] [PubMed] [Google Scholar]
- 8.Liljeström P., Lusa,S., Huylebroeck,D. and Garoff,H. (1991) In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol., 65, 4107–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 10.Sleat D.E., Gallie,D.R., Watts,J.W., Deom,C.M., Turner,P.C., Beachy,R.N. and Wilson,T.M.A. (1988) Selective recovery of foreign gene transcripts as virus-like particles in TMV-infected transgenic tobaccos. Nucleic Acids Res., 16, 3127–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss J.H. and Strauss,E.G. (1994) The alphaviruses: gene expression, replication and evolution. Microbiol. Rev., 58, 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong C., Levis,R., Shen,P., Schlesinger,S., Rice,C.M. and Huang,H.V. (1989) Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science, 243, 1188–1191. [DOI] [PubMed] [Google Scholar]
- 13.Levis R., Schlesinger,S. and Huang,H.V. (1990) Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J. Virol., 64, 1726–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garoff H., Huylebroeck,D., Robinson,A., Tillman,U. and Liljeström,P. (1990) The signal sequence of the p62 protein of Semliki Forest virus is involved in initiation but not in completing chain translocation. J. Cell Biol., 111, 867–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulvey M. and Brown,D.T. (1996) Assembly of the Sindbis virus spike protein complex. Virology, 219, 125–132. [DOI] [PubMed] [Google Scholar]
- 16.Lobigs M., Hongxing,Z. and Garoff,H. (1990) Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J. Virol., 64, 4346–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy R. and Huang,H.V. (1991) Analysis of Sindbis virus promoter recognition in vivo, using novel vectors with two subgenomic mRNA promoters. J. Virol., 65, 2501–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolov I. and Schelsinger,S. (1994) Translation of Sindbis virus mRNA: effects of sequences downstream of the initiation codon. J. Virol., 68, 8111–8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser C.A., Preuss,D., Grisafi,P. and Botstein,D. (1987) Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science, 235, 312–327. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y.F., Sawicki,S.G. and Sawicki,D.L. (1991) Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol., 65, 985–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs K.A., Collins-Racie,L.A., Colbert,M., Duckett,M., Golden-Fleet,M., Kelleher,K., Kriz,R., LaVallie,E.R., Merberg,D., Spaulding,V., Stover,J., Williamson,M.J. and McCoy,J.M. (1997) A genetic selection for isolating cDNAs encoding secreted proteins. Gene, 198, 289–296. [DOI] [PubMed] [Google Scholar]
- 22.Lim S.P. and Garzino-Demo,A. (2000) Cloning trap for signal peptide sequences. Biotechniques, 28, 124–130. [DOI] [PubMed] [Google Scholar]
- 23.Peterfy M., Gyuris,T. and Takacs,L. (2000) Signal-exon trap: a novel method for the identification of signal sequences from genomic DNA. Nucleic Acids Res., 28, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladunga I. (2000) Large-scale predictions of secretory proteins from mammalian genomic and EST sequences. Curr. Opin. Biotechnol., 11, 13–18. [DOI] [PubMed] [Google Scholar]
- 25.Kitamura T., Onishi,M., Kinoshita,S., Shibuya,A., Miyajima,A. and Nolan,G.P. (1995) Efficient screening of retroviral cDNA expression libraries. Proc. Natl Acad. Sci. USA, 92, 9146–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koller D., Ruedl,C., Loetscher,M., Vlach,J., Oehen,S., Oertle,K., Schirinzi,M., Deneuve,E., Moser,R., Kopf,M., Bailey,J.E., Renner,W. and Bachmann,M.F. (2001) A high-throughput alphavirus-based expression cloning system for mammalian cells. Nat. Biotechnol., 19, 851–855. [DOI] [PubMed] [Google Scholar]
- 27.Wielgosz M.M., Raju,R. and Huang,H.V. (2001) Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells. J. Virol., 75, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchholz C.J., Peng,K.W., Morling,F.J., Zhang,J., Cosset,F.L. and Russell,S.J. (1999) In vivo selection of protease cleavage sites from retrovirus display libraries. Nat. Biotechnol., 16, 951–954. [DOI] [PubMed] [Google Scholar]
- 29.Filocamo G., Pacini,L. and Migliaccio,G. (1997) Chimeric Sindbis viruses dependent on the NS3 protease of hepatitis C virus. J. Virol., 71, 1417–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pacini L., Vitelli,A., Filocamo,G., Bartholomew,L., Brunetti,M., Tramontano,A., Steinkühler,C. and Migliaccio,G. (2000) In vivo selection of protease cleavage sites by using chimeric Sindbis virus libraries. J. Virol., 74, 10563–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]