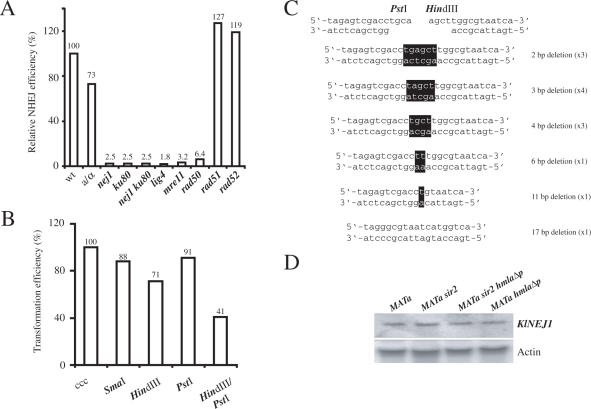
Figure 1.
K.lactis NHEJ was versatile and cell-type independent. (A) Molecular requirements for K.lactis NHEJ measured in a plasmid re-circularization assay. Strains CK213-4c (wt), SAY681 (MATa/MATα), SAY572 (nej1::LEU2), SAY573 (ku80::LEU2), SAY574 (nej1::LEU2 ku80::LEU2), SAY683 (lig4::KANMX), SAY559 (mre11::LEU2), SAY557 (rad50::KANMX), SAY516 (rad51::LEU2) and SAY507 (rad52::LEU2) were tested for end-joining efficiency by parallel transformation of super-coiled or HindIII-linearized pCXJ18. The data are expressed as the relative ratio between the number of transformants obtained with the linearized and the super-coiled plasmid, normalized to the value obtained for the wild-type strain that was defined as 100%. (B) Versatility of K.lactis NHEJ. Wild-type strain CK213-4c transformed with linearized pCXJ18 bearing blunt (SmaI), 5′ cohesive (HindIII) 3′ cohesive (PstI) or noncohesive (HindIII + PstI) overhangs were analyzed for transformation efficiency. The results represent the transformation efficiency obtained with the linearized plasmid relative to the efficiency obtained with super-coiled (ccc) pCXJ18, defined as 100%. The data shown in (A) and (B) correspond to the average value from two or three independent experiments. (C) Repair events from NHEJ of noncohesive overhangs. Plasmids were rescued from 13 independent K.lactis colonies transformed with HindIII/PstI double digested pCXJ18. The DNA sequences of the junctions (black boxes indicating the bases remaining from the overhangs) were determined, and the type of alteration and how many times they were obtained is indicated on the right. (D) RNA blot hybridization of K.lactis total RNA from CK213-4c (MATa), SAY102 (MATa sir2), SAY189 (MATa sir2 hmlαΔp) and SAY186 (MATa hmlαΔp) strains. The blot was hybridized with a 500 bp [α32P]-labeled PCR fragment corresponding to NEJ1 gene (top), and then the blot was stripped and re-probed with an actin probe (bottom).