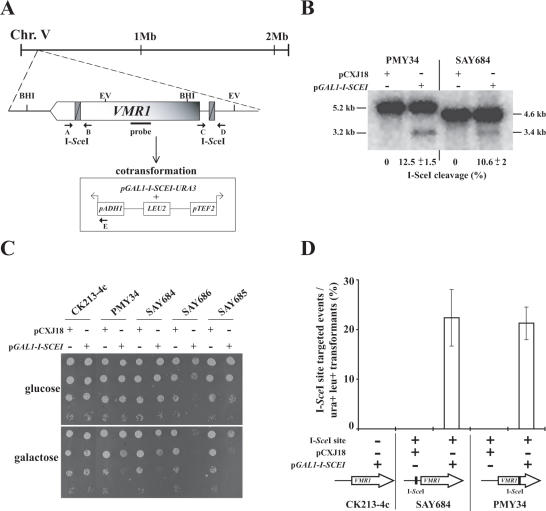
Figure 5.
Genomic DSBs constituted the target sites for IR events. (A) Schematic illustration of the VMR1 chromosomal locus. The I-SceI recognition site was introduced either in the VMR1 coding region (at nucleotide +3871, strain PMY34) or in its promoter region (at nucleotide −183, strain SAY684). In the experiment described in (D), cells were simultaneously transformed with an episomic plasmid containing the I-SCEI endonuclease under the GAL1 promoter (p164) and the BamHI-linearized pPMB18 that contains the ADH1 and TEF2 promoters, which directs transcription in opposite directions (indicated by arrows). The position of oligonucleotides (A, B, C, D and E) used for PCR analysis of the location of IR integration events and relevant restriction sites (BHI, BamHI; EV, EcoRV) are indicated. (B) The I-SceI endonuclease cleaves in the VMR1 coding sequence and in its promoter region with similar efficiency. Exponentially growing PMY34 and SAY684 cells containing either pCXJ18 or p164 in SC-Ura-glucose were washed, resuspended in SC-Ura-galactose and incubated for 3 h. Genomic DNA from PMY34 and SAY684 cells digested with BamHI or EcoRV, respectively, was separated, blotted and the membrane was hybridized with a 273 bp [α32P]-labeled PCR fragment corresponding to VMR1 gene as shown in (A). The fragment sizes are indicated on both sides of the blot and the I-SceI cutting efficiency is indicated below the blot with a standard deviation from two independent experiments. (C) Genomic DNA cleavage at the ectopic I-SceI sites results in a slow growth phenotype and is highly deleterious in combination with nej1 and rad52 mutations. Ten-fold serial dilutions of the indicated strains containing either pCXJ18 or p164 were spotted on SC-Ura containing either glucose or galactose as a carbon source and incubated at 30°C for 48 h. The relevant genotypes are: CK213-4c (WT), PMY34 (VMR1 +3871 I-SceI), SAY684 (pVMR1 −189 I-SceI), SAY686 (pVMR1 −189 I-SceI rad52) and SAY685 (pVMR1 −189 I-SceI nej1). (D) Strains CK213-4c, SAY684 and PMY34 were co-transformed either with pCXJ18 or p164 and BamHI-linearized pPMB18. Transformants were selected on SC-Ura-Leu containing galactose as the carbon source. IR insertion events of pPMB18 at both I-SceI sites were analyzed by PCR using the oligonucleotides indicated in (A). An additional phenotypic selection was carried out in those strains having the I-SceI site in the VMR1 promoter since insertion of a strong promoter in front of the VMR1 gene results in G418 resistance. The results are expressed as the percent of I-SceI site targeted events with respect to the total number of Ura+ Leu+ transformants and correspond to the average of three independent experiments. For control experiments 8 to 12 transformants were PCR analyzed. The sample size analyzed by PCR corresponding to the co-transformation of pPMB18 and p164 into PMY34 and SAY684 was 77 and 39, respectively. Phenotypic analysis for G418 resistance was performed with approximately 150 transformants in each case.