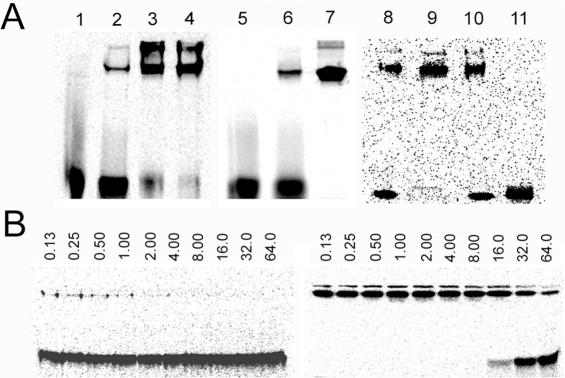
Figure 2.
The influence of B.subtilis SSB phosphorylation on ssDNA binding. (A) Gel shift assay with 0.2 pmol of [33P]labeled ssDNA (present in all lanes) with different quantities of B.subtilis SSB. Lane 1 contained only the ssDNA; lanes 2–4 contained 1, 10 and 100 pmol of SSB, respectively; lane 5 contained YwqD, YwqC-NCter, 1 mM MgCl2 and 5 mM ATP (negative control); lane 6 contained 2 pmol of SSB; lane 7 contained 2 pmol of SSB pretreated with YwqD, YwqC-NCter, 1 mM MgCl2 and 5 mM ATP for 1 h before the gel-shift. Lane 8 contained 5 pmol of SSB; in lane 9 the same quantity of SSB was in vitro phosphorylated (same treatment as in lane 7), and the sample from lane 9 was dephosphorylated by YwqE in the presence of 1 mM MnCl2 for 20 min (lane 10) and 60 min (lane 11). (B) Gel shift assay with SSB purified from SPSSBHT-ΔywqD. All reactions contained 125 nM SSB, and the total ssDNA concentration in nM (same amount of radiolabeled and increasing amount of unlabelled probe) is indicated above each lane. Left panel: unphosphorylated SSB, right panel: SSB phosphorylated in vitro before the gel shift study.