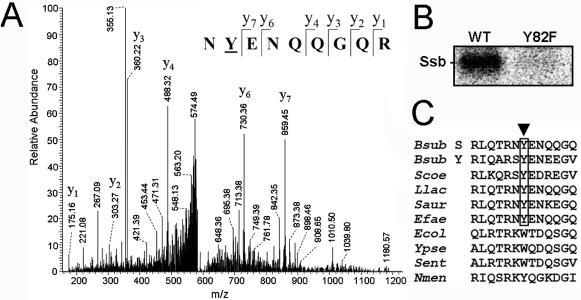
Figure 4.
Identification of the tyrosine phosphorylated residue in B.subtilis SSB. (A) MS/MS spectrum of the precursor ion at m/z 608.748, corresponding to the B.subtilis SSB peptide NYENQQGQR with one phosphorylated residue. C-terminal y-ion series confirms the peptide sequence and indicates Y82 as the phosphorylation site. The low intensity of the spectrum reflects low occupancy of the phosphorylation site. (B) In vitro phosphorylation of mutant protein BsSSB Y82F compared with wild-type BsSSB. Reactions were performed with YwqD-123. (C) Sequence alignment of the region surrounding the phosphorylated residue Y82 in B.subtilis SSB. Aligned bacterial SSBs are from B.subtilis (Bsub), S.coelicolor (Scoe), Lactococcus lactis (Llac), Staphylococcus aureus (Saur) Enterococcus faecalis (Efae), Escherichia coli (Ecol), Yersinia pseudotuberculosis (Ypse) Salmonella enterica (Sent) and Neisseria meningitidis (Nmen). B.subtilis SSB and YwpH are denoted by ‘S’ and ‘Y’, respectively.