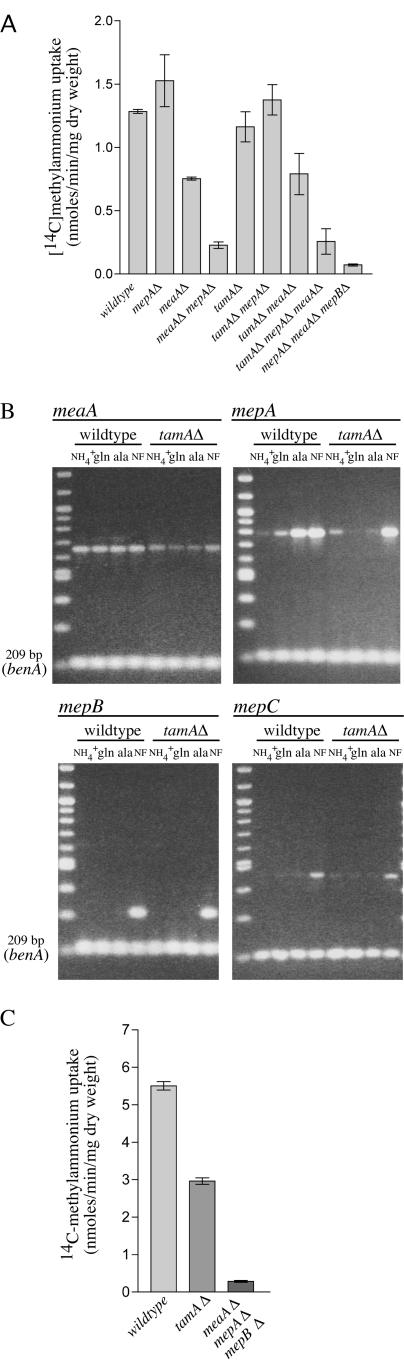
FIG.7.
Analysis of A. nidulans AMT/MEP genes in tamAΔ mutant background. (A) [14C]methylammonium transport rates forwild-type (MH1) and mepAΔ (MH9831), meaAΔ (MH9830), meaAΔ mepAΔ (MH9829), tamAΔ (MH8694), tamAΔ mepAΔ (MH10316), tamAΔ meaAΔ (MH10315), tamAΔ meaAΔ mepAΔ (MH10317), and meaAΔ mepAΔ mepBΔ (MH10325) mutant strains. Assays were performed with nitrogen-starved mycelia at a final methylammonium concentration of 200 μM. (B) RT-PCR analysis of meaA, mepA, mepB, and mepC expression in wild-type and tamAΔ mutant strains. RNAs were isolated from mycelia grown for 16 h at 37°C in 1% glucose medium, pH 6.5, with 20 mM ammonium (NH4+) and then transferred to nitrogen-free (NF) medium, pH 6.5, for 4 hours or from mycelia grown for 16 h at 37°C in 1% glucose medium, pH 6.5, with 10 mM glutamine (gln) or alanine (ala), as indicated. The name of the respective gene is shown at the top of each picture, and in all cases, the lower band is benA. (C) [14C]methylammonium transport rates for the wild-type and tamAΔ and meaAΔ mepAΔ mepBΔ mutant strains for mycelia grown for 16 h with 10 mM alanine. A final methylammonium concentration of 200 μM was used. Error bars represent standard errors from at least two independent experiments.