Figure 5. Structural distinctions between prion strain populations.
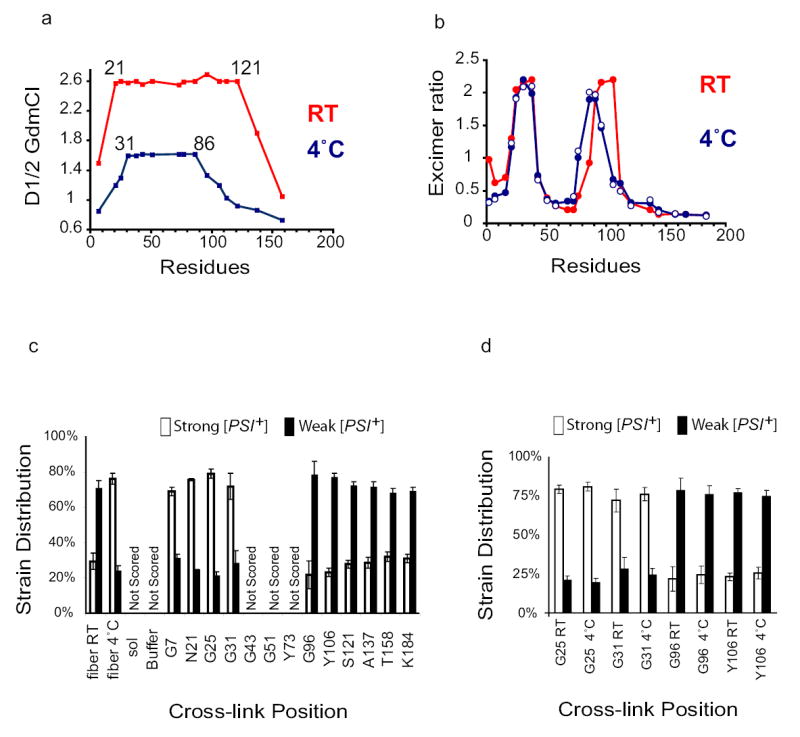
a) Amyloid core length and stability differ in fibres assembled at 4°C and 25°C. Acrylodan-labelled fibres assembled at 4°C ( ) and 25°C (
) and 25°C ( ) were denatured as in Fig. 1b. D1/2 values were obtained from full GdmCl denaturation profiles (see Fig 1c).
) were denatured as in Fig. 1b. D1/2 values were obtained from full GdmCl denaturation profiles (see Fig 1c).
(b) Intersubunit interfaces change in fibres assembled at different temperatures. Excimer fluorescence of pyrene-labelled mutants assembled in rotated unseeded reactions at 25°C ( ), 4°C (
), 4°C ( or in seeded unrotated reactions with 4% seed formed at 4°C (
or in seeded unrotated reactions with 4% seed formed at 4°C ( ). Pyrene fluorescence is similar in seeded and unseeded reactions.
). Pyrene fluorescence is similar in seeded and unseeded reactions.
(c) BMB cross-links at different positions bias assembly towards distinct prion strains. NM proteins cross-linked in denaturant were diluted into buffer, assembled into fibres, and used to transform [psi−] cells to [PSI+] state. Left, controls: uncross-linked WT fibres assembled at RT or at 4°C are biased towards the production of weak or strong strains, respectively27. Soluble NM, buffer alone, and proteins cross-linked in the Central Core did not induce [PSI+] above background and were not scored. Values represent means ± SD (n=5 experiments)
(d) Strain biases created by cross-linking overcome the biases created by assembly at different temperatures. Whether they were assembled at 4°C or at RT, fibres cross-linked in the Head (G25C or G31C) produced mostly strong [PSI+] strains, and fibres cross-linked in the Tail (G96C and106C) produced mostly weak [PSI+] strains. Values represent means ± SD (n=5 experiments)
