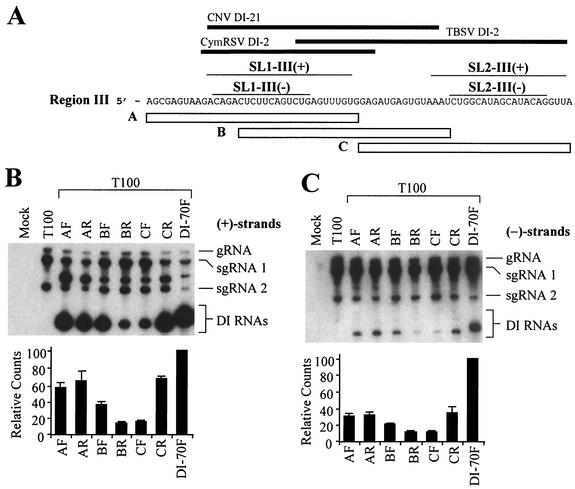
FIG. 5.
Analysis of DI RNAs containing three different overlapping 35-nucleotide segments of region III. (A) The plus strand sequence of region III is presented. Open bars below the sequence represent segments A, B, and C. Thick black lines at the top represent corresponding region III segments present in cucumber necrosis virus (CNV) DI-21, cymbidium ringspot virus (CymRSV) DI-2, and TBSV DI-2 (10, 14, 17), and the thin lines directly below show corresponding positions of SL1-III(+), SL1-III(−), SL2-III(+), and SL2-III(−). (B) Northern blot analysis of plus strands of DI RNAs containing region III segments (outlined in Fig. 4A) introduced in either the forward (F) or reverse (R) orientation. Approximately 4.0 × 105 cucumber protoplasts were inoculated with H2O (mock) or 2 μg of T100 transcript only or coinfected with 2 μg of T100 and 1 μg of the respective DI RNAs. Total nucleic acids were isolated 24 h postinoculation, separated in 8 M urea-4.5% polyacrylamide gels, transferred to nylon membranes, and subsequently probed with a 32P-end-labeled minus-sense probe. Bands corresponding to TBSV genomic RNA (gRNA), subgenomic mRNA1 (sgRNA1), subgenomic mRNA2 (sgRNA2), and various DI RNAs are shown. The bands between sgRNA1 and sgRNA2 in coinfections represent previously described head-to-tail dimers of DI RNAs (32). Quantification of DI RNA bands (monomers only) is shown in the bar graph below the autoradiogram in panel B. (C) Northern blot analysis of corresponding minus strands for the samples in B. Minus strands were analyzed as in panel B except that membranes were hybridized with plussense probes.