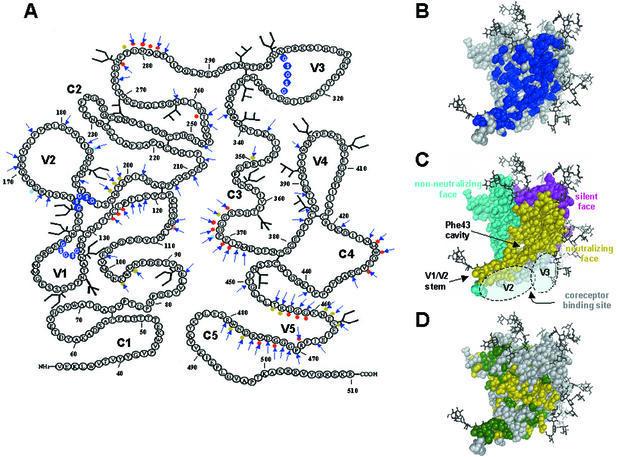
FIG. 1.
Locations of amino acid substitutions on gp120, spatial relationships of epitopes on gp120, and variability of gp120 residues among primate immunodeficiency viruses and HIV-1. (A) Ribbon amino acid sequence diagram of gp120JR-CSF indicating the locations of the alanine substitutions in this study. Arrows indicate amino acids that were mutated to alanine. Variable-loop deletions are indicated by white letters encircled by blue spheres. Light blue dots indicate the positions of amino acid mutations in b12 neutralization escape mutants (38, 54). Red and yellow dots indicate primary and secondary b12 contact residues, respectively, based on a computational docking model of b12 and the gp120HxB2 core (44). Primary b12 contacts are gp120 residues that, based on our docking model, contact MAb b12; secondary contacts are gp120 residues within 3 to 5 Å of MAb b12. (B) Locations of mutations (shown in blue) mapped onto the structure of the gp120 core of HIV-1HxB2. The view is from the perspective of CD4. (C) Approximate locations of the faces of the gp120 core, defined by the interactions of gp120 and antibodies (30, 76, 78). The region in the CD4bs that is accessible to neutralizing ligands on primary HIV-1 isolates (i.e., MAbs CD4-IgG2 and b12), termed the neutralizing face, is shown in yellow. The region that is believed to be poorly accessible on oligomeric gp120 and that elicits nonneutralizing antibodies is shown in cyan. The location of the immunologically silent face, which encompasses the epitope recognized by broadly neutralizing MAb 2G12 (59, 62, 73), is shown in magenta. The coreceptor binding site is shown in light grey. Modeled carbohydrate chains are shown in dark grey and black. The approximate areas that are believed to be covered by the V2 and V3 loops (primarily the coreceptor binding site) are indicated. The locations of the Phe43 cavity (30), involved in CD4 binding, and the stem of the V1/V2 loop are also indicated. (D) Molecular surface of gp120 depicting the sequence variability of the amino acid residues among primate immunodeficiency viruses and HIV-1: green, residues that are conserved among all primate immunodeficiency viruses; yellow, residues that are conserved among all HIV-1 isolates but not among all primate immunodeficiency viruses; grey, residues that are variable among HIV-1 isolates. Amino acid conservation is defined as in reference 30.