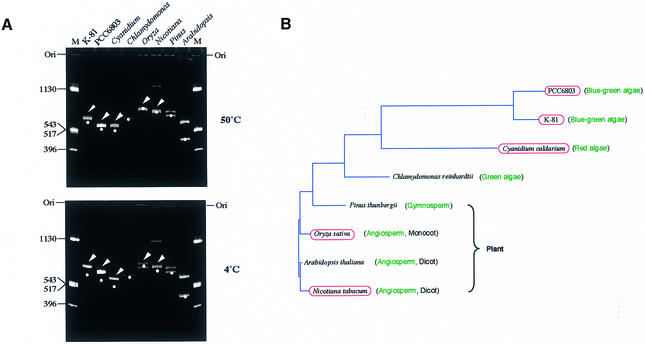
Figure 2.
Universality of intrinsic curvatures in the 5′-upstream regions of the psbA family. (A) Polyacrylamide gel electrophoresis (6%) at 4 and 50°C was performed with psbA fragments prepared as follows: M.aeruginosa K-81 psbA2 (–404/+111, transcription start point as +1), a 639 bp PCR fragment amplified by M4 and RV primers (these primers were used to amplify an insert DNA cloned at the multiple cloning site in pUC derivatives) with pHNL7-up (4); Synechocystis PCC 6803 psbA2 (–451/+79, transcription start point as +1), a 629 bp PCR fragment amplified by the M4 and RV primers with pSBA2 (Amano et al., unpublished results); C.caldarium psbA (–474/+24, A of the start codon ATG as +1), a 498 bp PCR fragment amplified by PSBA-Cyan-F and PSBA-Cyan-R primers with the genomic DNA; Chlamydomonas reinhardtii psbA (an ∼1000 bp upstream region), pR14 (38) was digested with HindIII, isolated as a small segment, then the fragment digested with DdeI (635 bp HindIII–DdeI and 365 bp DdeI–HindIII fragments appeared); Pinus thunbergii psbA (a 1.3 kb BamHI–PstI fragment, –1040/+260, A of the start codon ATG as +1), a PCR fragment, amplified by the M4 and RV primers with pSBA-B (a pUC progenitor, Matsuo et al., unpublished results), was digested with AseI (707 and 656 bp fragments appeared); O.sativa psbA (a 1.6 kb BamHI–PvuII fragment, –1100/+460, A of the start codon ATG as +1), a PCR fragment amplified by the M4 and RV primers with pSBA-R (a pUC progenitor, Matsuo et al., unpublished results), was digested with AccI (770 and 900 bp fragments appeared); Arabidopsis thaliana psbA (a 1.0 kb fragment, –960/+35, A of the start codon ATG as +1), an EcoRV–BglII PCR fragment amplified by AraPSBA-F2 and AraPSBA-R primers with pMAH2 (39) was digested with AccI (550 and 450 bp fragments appeared); N.tabacum psbA (a 1.37 kb BamHI–PstI fragment, –1100/+267, A of the start codon ATG as +1), a PCR fragment amplified by the M4 and RV primers with pSBA-T (a pUC progenitor, Matsuo et al., unpublished results) was digested with DraI (741 and 731 bp fragments appeared; a small amount of the undigested PCR fragment was also observed as contamination). The positions of moleculer size markers (pUC119 DNA digested by HinfI) are indicated in lane M. Bands shown by arrowheads contain intrinsic DNA curvature. Dots indicate the position corresponding to the expected size of each psbA fragment. Positions of the gel origin (Ori) are presented. (B) A phylogenetic tree of amino acid sequences of the D1 (PsbA) proteins and conservation of the DNA curvature. The red ellipses indicate strains containing an intrinsic curvature in the region 5′-upstream of the psbA gene. The phylogenetic tree was calculated by the neighbor joining method (GENETYX-MAC software; Software Development Co., Japan).