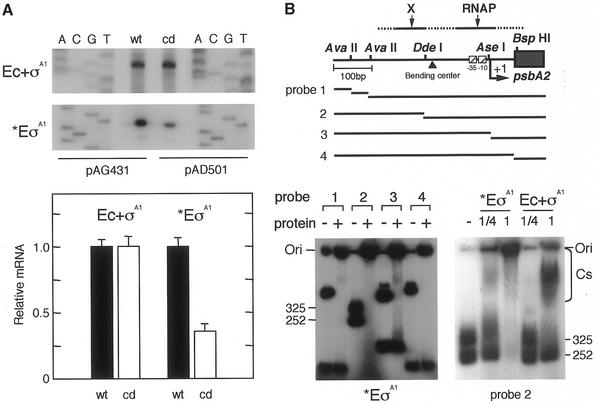
Figure 5.
In vitro psbA2 basal transcription and factors contributing to the transcription. (A) In vitro mRNA synthesis by two different RNAPs from the mutagenized CIT promoter. Using the RNAP of Ec + σA1 (1 pmol purified E.coli core enzyme + 3 pmol K-81 principal sigma factor σA1) or *EσA1 [partially purified RNAP fraction containing K-81 σA1; 15 µl (∼2 pmol RNAP)], in vitro mRNA synthesis was carried out from the DNA template pAG431 or pAD501 (3 µg). The synthesized mRNA was examined by in vitro primer extension with the primer lacZ-R1 (7). Relative values of synthesized mRNA (%) according to the signal intensity on X-ray film are shown. (B) DNA binding of RNAP components to the psbA2 upstream region. (Top) A schematic representation of template DNAs. A 576 bp EcoRI (E)–HindIII (H) fragment (Fig. 1) isolated from pHNL7-up (the –404 to +113 psbA2 region) (7) was digested with the respective restriction enzymes (see Fig. 1). These fragments were end-labeled, then the mixture was subjected to the assay. DNA fragments (1 µl, 1 pmol) were mixed with (left) K-81 *EσA1 (0.25 or 1 µl RNAP at 0.4 pmol/µl) or (right) Ec + σA1 (0.25 or 1 µl at 0.4 pmol/µl). The gel profile for no addition of protein is indicated as –. The positions of the DNA fragments, DNA–protein complexes (Cs) and gel origin (Ori) on the gel are shown.