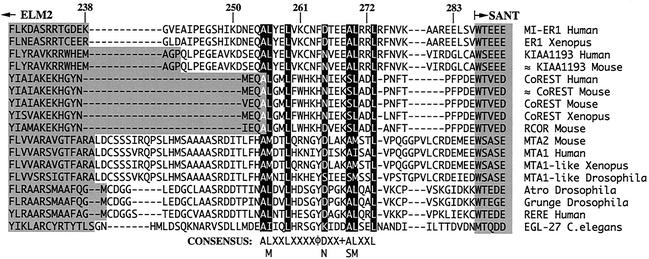
FIG. 6.
Alignment of ELM2 domains reveals additional conserved sequence. The ELM2 regions of proteins from the Pfam and GenBank databases were aligned using ClustalW. Shown is the amino acid sequence from the C-terminal end of the ELM2 domain to the beginning of the SANT domain in each protein. Residues belonging to these two domains are shaded. Highly conserved residues in the region C terminal to the ELM2 domain are shown with white lettering and highlighted in black. The consensus sequence is listed below the alignment; X represents any amino acid; φ represents Y, F, or H; and + represents a charged residue. The numbers listed above the alignment correspond to amino acid positions in the hMI-ER1 protein sequence. The accession numbers for the sequences used in this alignment (from top to bottom) are as follows: AF515447, O42194, AB033019, XM_125783, Q9UKL0, XM_127140, XM_127140, AJ311849, Q9JMK4, Q9R190, Q13330, O94776, Q9VNF6, Q9VNF6, Q9NHX6, Q9P2R6, and Q09228.