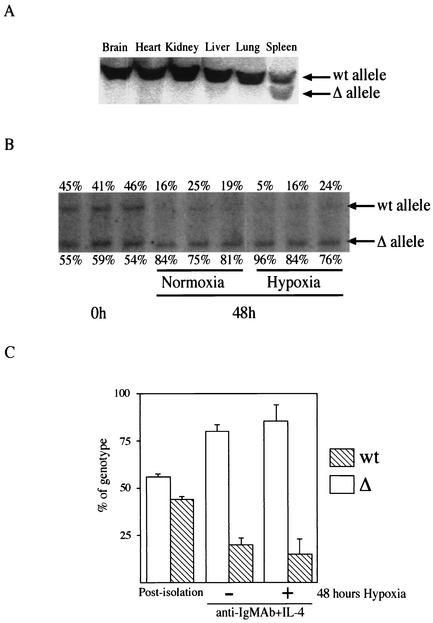
FIG. 1.
Deletion of the HIF-1α allele in B cells in vivo and in vitro. (A) Tissue-specific deletion of HIF-1α caused by CD19cre expression. Southern blot analysis of DNA from different organs of HIF-1α null (HIFDFCD19cre) mice was performed. DNA was digested with EcoRI and PstI and probed with a fragment of HIF-1α intron 1 as described elsewhere (27). The positions of DNA with the HIF-1α floxed allele (wt) and DNA in which the HIF-1α allele had been deleted (Δ) are shown to the right of the blot. (B) Increased HIF-1α-deficient B cells in response to mitogen. B cells isolated from HIF-1α null (HIFDFCD19cre) mice were stimulated with anti-IgM antibody (Ab) (10 μg/ml) plus IL-4 (0.5 ng/ml) for 48 h under either normoxia or hypoxia. After the stimulation, DNA was extracted and analyzed as described above for panel A. The data shown are representative results from three different preparations. The values immediately above and below the blot are the percentages of wild-type B cells and HIF-1α null cells, respectively. Note that HIF-1α null cells expanded preferentially in response to mitogen under both normoxia and hypoxia compared to wild-type B cells in culture. (C) Quantification of the proliferative advantage of HIF-1α null B cells in culture. The histogram is a graphic representation of information from panel B. Error bars show standard errors.