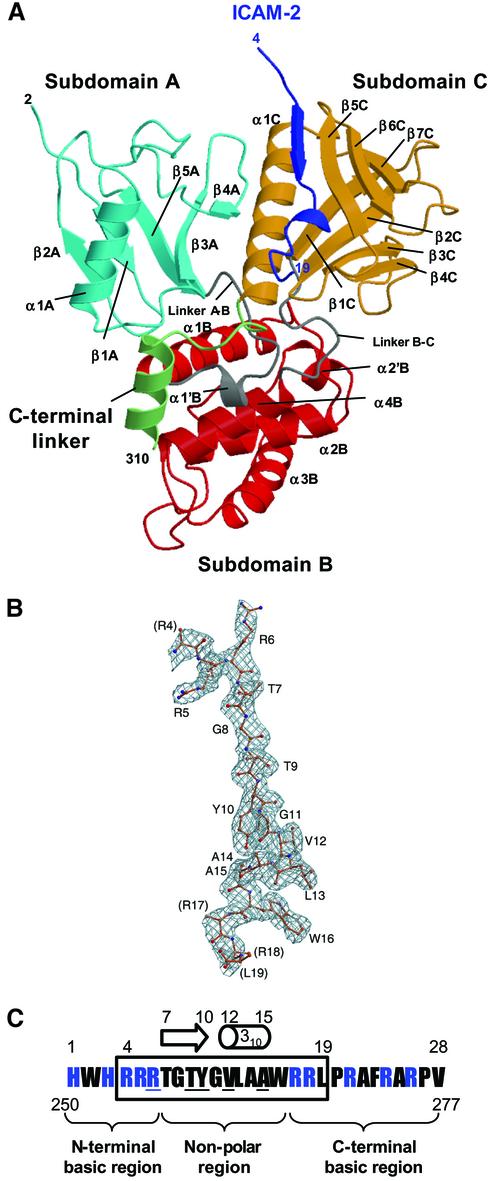
Fig. 1. Overall structure of the radixin FERM domain bound to the ICAM-2 tail peptide. (A) Views of the radixin FERM domain bound to the ICAM-2 peptide by ribbon representations. The ICAM-2 peptide is shown in blue. The radixin FERM domain consists of subdomains A (light blue), B (red) and C (brown). The linkers A–B (residues 83–95) and B–C (residues 196–203) are colored in gray and the C-terminal linker in green. (B) The ICAM-2 peptide model in a 2Fo–Fc electron density map countered at the 1σ level. The amino acid residues are indicated with labels of one-letter codes. Labels in parentheses indicate the terminal residues whose side chains were not defined in the map. (C) The 28-residue peptide synthesized based on the sequence of the mouse ICAM-2 cytoplasmic tail was used for the structural work. Basic residues are in blue. This peptide has two basic regions and a non-polar region between them. The 16 residues of the peptide defined on the current map are boxed. Key residues in binding to the radixin FERM domain are underlined (see text). The short β-strand (residues 7–10) and one 310 helix (resides 12–15) are indicated with an arrow and a cylinder.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.