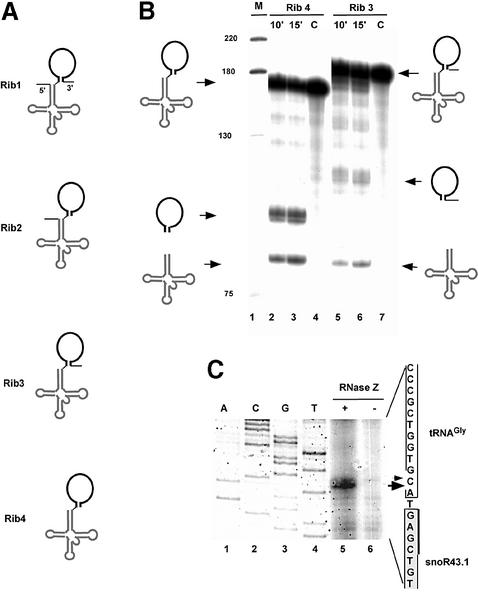
Fig. 5. Cleavage of the tRNA–snoRNA precursor by recombinant RNase Z in vitro. (A) Structures of RNA substrates used for processing. All substrates contain tRNA (grey cloverleaf) and snoRNA (black stem loop). Substrates differ in the presence of the 5′ leader and 3′ trailer sequences shown by thin lines. (B) Processing of RNA substrates Rib3 and Rib4. Precursor RNAs Rib4 (5′ and 3′ end matured) and Rib3 (5′ matured) were incubated with rRNase Z for 10 min and 15 min as indicated. C are control reactions performed without addition of proteins. M is the DNA size marker given in nucleotides on the left. Precursor and products are shown schematically at the sides. (C) Primer extension analysis of the rRNase Z cleavage site on Rib4 substrate. Unlabelled Rib4 substrate was incubated in vitro with or without rRNase Z as indicated. After incubation, products of the reaction were extracted and analysed by primer extension with a radiolabelled primer snoR4 annealing to the snoRNA. ACGT refers to sequencing reactions started from primer sno4. The position of the cleavage site on the coding strand is indicated with an arrow. The small arrow indicates a minor cleavage site.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.