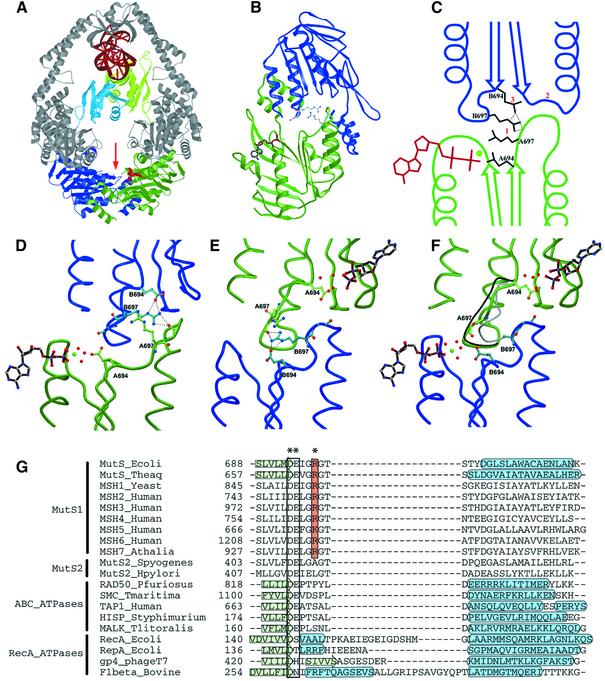
Fig. 6. Structural analysis of asymmetry. (A) Structure of the MutS dimer in complex with mismatched DNA. Protein is coloured in grey, with ATPase domains of monomer A and B coloured in green and blue, respectively, and mismatch binding domains of monomer A and B in light green and light blue. DNA and ADP are in red. (B) The two ATPase domains viewed along the arrow in (A). The ADP molecule in monomer A is coloured in brown; the side chains of the two Arg697 (coloured in grey) are located at the centre of the interface of the two ATPase domains. (C) Schematic representation of the asymmetric interactions of the ATPase domains of monomer A (green) and B (blue). (1) ArgB697 hydrogen-bonds to the backbone of GlyA698 in the DE-loop of monomer A, while the reverse contact does not take place. (2) As a result, the DE-loop of monomer A clashes with the P-loop of monomer B, which is thus inhibited from nucleotide binding. (3) Simultaneously, ArgB697 also hydrogen-bonds to and displaces GluB694. (D) Close-up of the nucleotide-binding site of monomer A in green, with the opposing monomer B in blue. (E) Same view as (D), but now viewed from monomer B. (F) Same view as (E), but of R697A MutS. The position of the DE-loop in monomer A and B of wild-type MutS is indicated in grey and black, respectively. In the absence of Arg697, no contacts are made between the two DE-loops, and both monomer A and B now bind ADP-Mg2+. (G) Structure-based sequence alignment of MutS homologues and paralogues, and related ATPases. The conserved Arg697 is coloured in orange, and marked with an asterisk (*). The Walker B motif is boxed, and indicated by (**). Secondary structural elements are indicated by a green arrow (β-strand) or a blue tube (α-helix). PDB accession codes E.coli MutS, 1E3M; Taq MutS, 1EWQ; RAD50, 1F2U; SMC, 1E69; TAP1, 1JJ7; HisP, 1B0U; MalK, 1G29; RecA, 2REB; RepA, 1G8Y; gp4, 1E0J; F1, 1BMF.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.