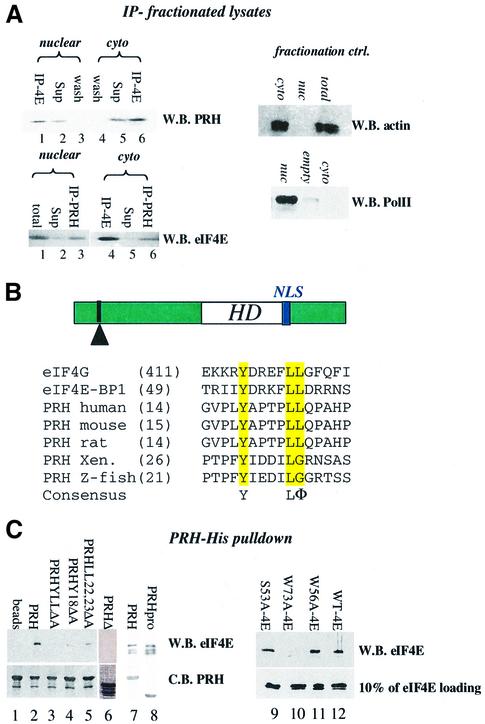
Fig. 2. PRH and eIF4E interact directly. (A) U937 cells were fraction ated, immunoprecipitated and analyzed by western analysis. IP refers to the immunoprecipitated fraction, Sup to supernatant after IP, and wash to last wash of protein A beads after IP. Fractionation controls are given in the adjacent panels, using actin as a cytoplasmic marker and RNA polymerase II as the nuclear marker. Total indicates total cell lysate; nuc, nuclear fraction; and cyto, cytoplasmic fraction. (B) Schematic of the PRH protein showing the position of the homeodomain (HD) relative to the eIF4E-binding site (indicated by the arrowhead) and the location of the NLS (in blue). The eIF4E-binding site in PRH from several organisms is shown. Accession numbers are as follows: NP-002720 (human), NP-032271 (mouse), NP-077361 (rat), AAB82335 (Xenopus) and NP-571009 (zebrafish). (C) His tag pull-down analysis of the PRH–eIF4E interaction. Wild-type (right panel) or mutant PRH proteins were immobilized on nickel–agarose beads and incubated with wild-type or mutant eIF4E as indicated. Loading of PRH mutants is shown in the bottom left panel. All constructs produced proteins at the expected molecular weight. Faster migrating bands indicate degradation products. eIF4E proteins used in lanes 7 and 8 are slightly degraded; the top band corresponds to full-length protein. Loading of eIF4E mutants is shown by western blot (W.B.) in the lower right panel. Equivalent amounts of eIF4E were used for experiments in the right panel where binding is ∼10% of eIF4E input. C.B. indicates Coomassie Blue-stained SDS–polyacrylamide gel.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.