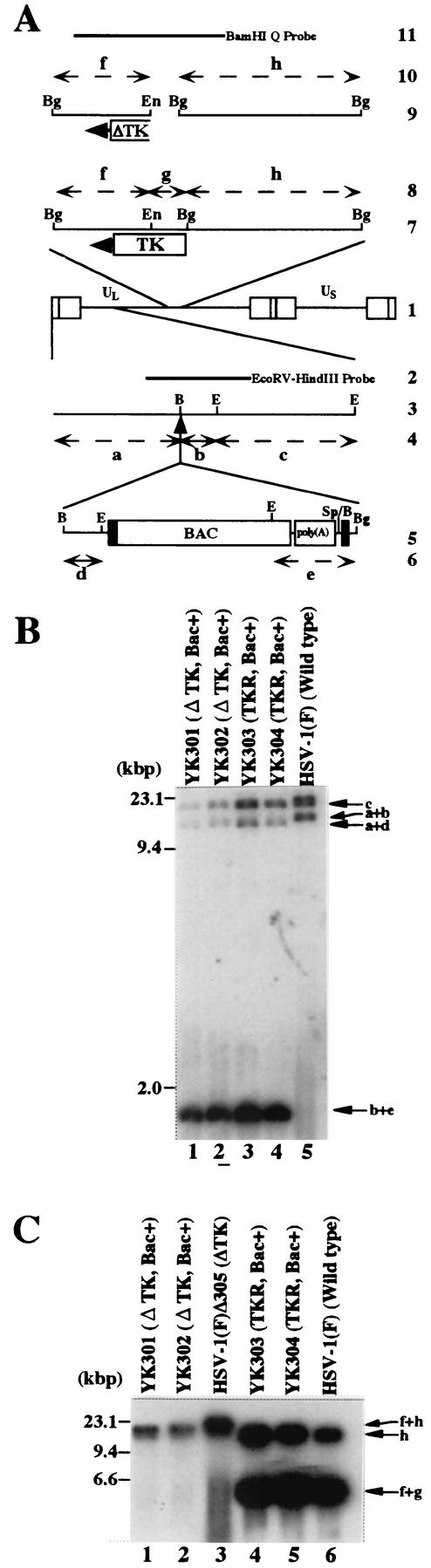
FIG.2.
(A) Schematic diagram of genome structures of HSV-1(F) and relevant domains of the recombinant viruses. Line 1, sequence arrangement of HSV-1(F) genome. Line 2, location of the EcoRV-HindIII fragment used as a radiolabeled probe in panel B. Line 3, an enlarged portion of the EcoRI J and D fragments of HSV-1(F). Line 5, sequence of the BAC-containing recombinant virus YK301, YK302, YK303, or YK304. Lines 4 and 6, expected sizes of DNA fragments generated by cleavage of the DNA. The fragment designations shown here are identical to those described in the text and in panel B. Line 7, an enlarged portion of the BglII M and I fragments of HSV-1(F), YK303 or YK304. Line 9, Δ305, YK301, or YK302 has a deletion (fragment g) in the TK locus. Lines 8 and 10, expected sizes of DNA fragments generated by cleavage of the DNA. The fragment designations shown here are identical to those described in the text and in panel C. Line 11, location of the BamHI Q fragment used as a radiolabeled probe in panel C. Restriction sites: E, EcoRI; B, BamHI; Bg, BglII; En, EcoNI; Sp, SphI. (B and C) Autoradiographic images of electrophoretically separated EcoRI (B) or BglII (C) digests of YK301, YK302, YK303, YK304, HSV-1(F)Δ305, and HSV-1(F) DNAs, hybridized to the radiolabeled EcoRV-HindIII fragment of pRB442 or BamHI Q fragment of HSV-1(F), respectively. The letters on the right refer to the designations of the DNA fragments generated by restriction endonuclease cleavage. Molecular sizes are shown on the left.