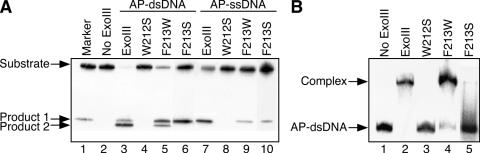
Figure 2.
Influence of substitutions at the conserved aromatic amino acid residues around the active site of ExoIII. (A) Detection of products of cleavage by ExoIII and its mutant proteins. Proteins used in each reaction are shown above each lane. The oligonucleotide containing an AP site (AP-ssDNA) was d-GCGATGACTAACGHTACTAGGCTTCCGAGCC. AP-dsDNA was constructed by annealing AP-ssDNA and d-CGGAAGCCTAGTATCGTTAGTCATCGCCATG. The substrate DNA (4.4 pmol), in which the oligonucleotide containing an AP site was 5′ 32P-labeled, was incubated with the wild-type ExoIII or its mutant (0.04 pmol) in buffer containing 75 mM NaCl, 5 mM MgCl2, 10 mM DTT and 65 mM Tris–HCl (pH 8.0) at 23°C. The DNA products were analyzed using a 20% denaturing (7 M urea) polyacrylamide gel. The product 1 was produced by AP endonucleolytic cleavage at the 5′ side of the AP site. The product 2 was produced by exonucleolytic cleavage from 3′ end of the product 1. The markers were 5′ 32P-labeled d-GCGATGACTAACG. (B) Abilities of ExoIII and its mutant proteins to bind the dsDNA containing an AP site. The proteins used in each reaction are shown above each lane. AP-dsDNA (0.45 pmol), in which the oligonucleotide containing an AP site was 5′ 32P-labeled, was incubated with the wild-type ExoIII or its mutant (4.5 pmol) in buffer containing 77 mM NaCl, 10 mM EDTA and 66 mM Tris–HCl (pH 8.3) at 4°C. The protein–DNA complex was analyzed by 15% native polyacrylamide gel electrophoresis.