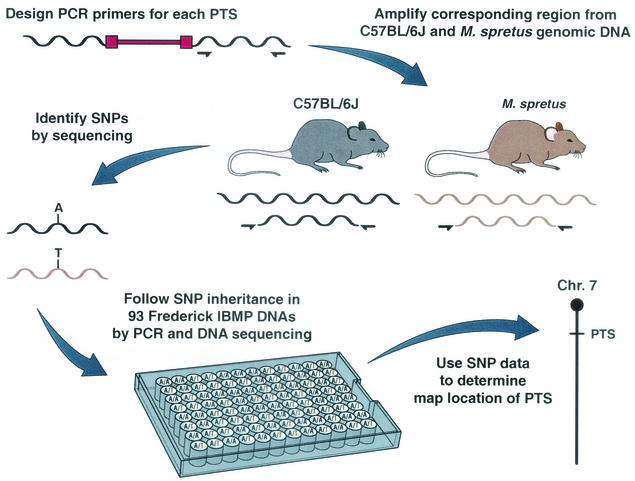
FIG. 1.
High-throughput, SNP-based mapping of PTSs cloned from mouse hematopoietic tumors. PCR primer pairs were generated for 240 noninformative PTSs (12) and used to amplify 250- to 350-bp fragments from C57BL/6J and M. spretus genomic DNA (10). Proviral DNA is shown in red, and the locations of amplification primers in the flanking cellular DNA are shown by half-arrows. Amplification reaction mixtures contained 4 ng of mouse DNA, 1 μg of forward and reverse primers, 2 U of Platinum Taq polymerase (Gibco BRL), and the buffer supplied by the manufacturer. To improve the specificity of the PCR, we used a touchdown PCR program for the first 12 cycles. The annealing temperature was decreased by 1°C from 65 to 60°C after every two cycles. This was followed by 25 cycles of 94°C for 20 s, 56°C for 20 s, and 68°C for 1 min and a final extension of 68°C for 5 min on a GeneAmp PCR System 9700 (PE Applied Biosystems). The amplification products were then sequenced to identify SNPs or insertion-deletion polymorphisms. PCR products were pretreated to remove excess primers and deoxynucleoside triphosphates before they were sequenced. Each sample was incubated with 1 U of shrimp alkaline phosphatase (Amersham)/μl and 10 U of exonuclease (Amersham)/μl at 37°C for 5 min, followed by 72°C for 15 min to denature the enzyme. The amplification products were sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing kit (PE Applied Biosystems) with the forward primer. Sequence files were analyzed with PolyPhred (Washington University) for simple SNPs or with MultiALN for polymorphisms involving insertions and deletions. Polymorphic primer pairs were then used to amplify 93 DNAs from the Frederick IBMP along with C57BL/6J and M. spretus control DNA. The 205 Frederick IBMP progeny were generated by mating (C57BL/6J × M. spretus)F1 females and C57BL/6J males as previously described (4). By monitoring the inheritance of these polymorphisms in the backcross DNAs, it was possible to accurately position each PTS on the Frederick IBMP.