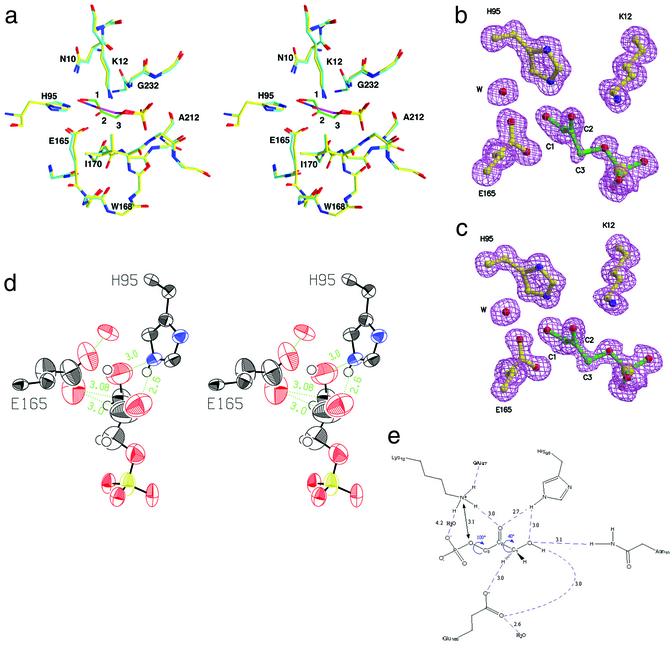
Figure 3.
Structure of yeast TIM:DHAP complex. (a) Triosephosphate isomerase in complex with a tight-binding transition-state analog phosphoglycolohydroxamate [PDB entry 7TIM (9) in cyan and purple for carbon atoms in the enzyme and the analog, respectively] is overlaid with our structure of the actual substrate, DHAP (in yellow and green). The position of the substrate differs in fine details from that of the transition-state analog. (b) Omit electron density for the substrate and catalytic residues Glu-165, His-95, and Lys-12. (c) Omit electron density for the active site in the second molecule. (d) ORTEP representation displaying the anisotropic motion for the active-site atoms. The glutamic acid side chain and the substrate carbons (C3, C2, and less so, C1) as well as the oxygens undergoing isomerization (O1 and O2) are more dynamically flexible than the active-site residues and phosphate group. (e) A schematic drawing of the network of hydrogen bonds in the active site.