FIG. 1.
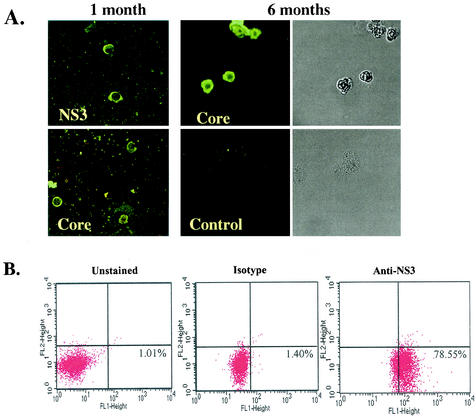
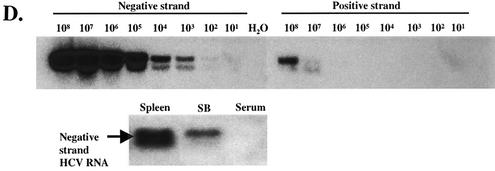
SB cells are persistently infected by HCV. (A) Detection of HCV protein expression by immunofluorescence staining in SB cells at different time points of culture. Uninfected Raji cells were used as a control. Phase contrast images of SB cells (upper) and uninfected Raji cells (lower) are shown on the right. (B) Detection of intracellular NS3 protein expression in SB cells by flow cytometry. Unstained SB cells served as the “no-antibody” control; isotype control antibody served as the background staining control. (C) Detection of positive-strand HCV RNA in SB cells by RPA. The expected size of the protected band from HCV RNA in SB cells is 123 bp. Different amounts of in vitro-transcribed sense and antisense HCV RNA (IVT-S and IVT-AS) were used for quantitation. The AS probe was used for detecting positive-strand RNA, and the S probe was used for detecting negative-strand RNA. Because of the presence of vector sequences in both the probes and the in vitro-transcribed RNA, the protected sizes for IVT-S and IVT-AS are 215 and 233 bp, respectively. HCV(+), SB RNA hybridized to the AS probe; HCV(-), same RNA hybridized to the S probe. (D) Detection of negative-strand HCV RNA by strand-specific RT-PCR and Southern blotting. The top blot shows the validation of the strand specificity of the assay. Serial dilutions of both positive- and negative-strand in vitro-transcribed HCV RNAs from 108 molecules to 1 molecule were subjected to negative-strand-specific RT-PCR using the sense primer for reverse transcription. The bottom blot shows that negative-strand HCV RNA was detected in the spleen and SB cells but not in the patient's serum.