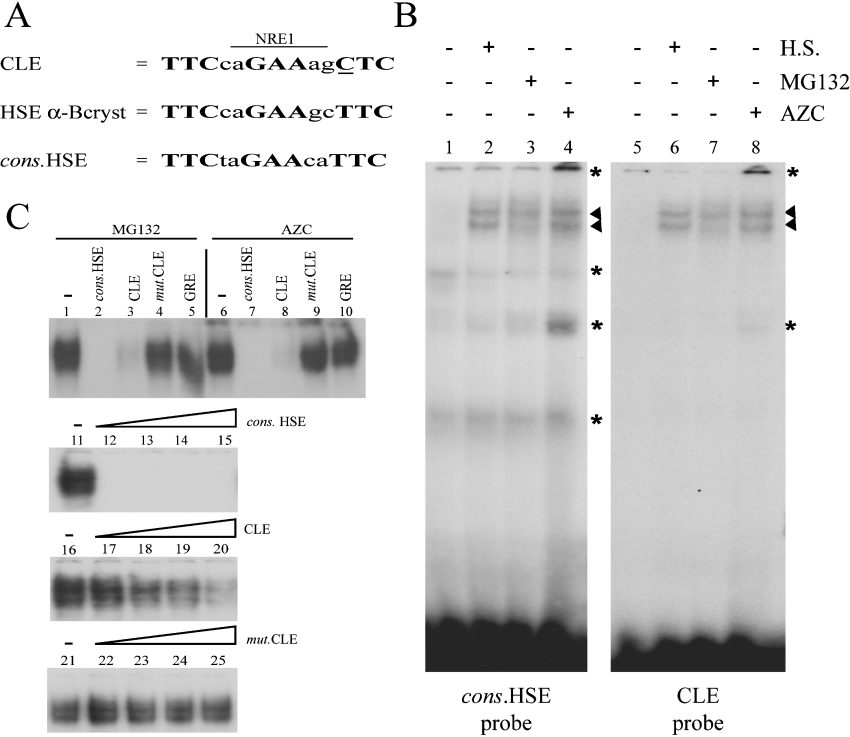
Figure 3. MG132 and AZC treatments increase protein DNA-binding activities on CLE.
(A) The conserved CLE sequence is a composite responsive element containing an NRE (NRE-1) overlapping a degenerate HSE. The three binding sites of HSEs are in boldface and the non-consensus nucleotide within CLE is underlined. The cons.HSE and αB-crystallin HSE are shown for comparison. (B) Gel-shift experiments with consensus HSE (lanes 1–4) or CLE (lanes 5–8) 32P-labelled double-stranded oligonucleotides as probes. The binding reaction was performed with cell extracts from U-251 MG control cells (lanes 1 and 5), 42 °C heat-shock (H.S.)-treated cells (lanes 2 and 6), 5 μM MG132-treated cells (lanes 3 and 7) and 25 μM AZC-treated cells (lanes 4 and 8). The position of the specific DNA–protein complexes are marked by arrowheads and the non-specific DNA–protein interactions by asterisks. (C) Competition experiments with 32P-labelled CLE oligonucleotides as probe and cell extracts from MG132- (lanes 1–5 and lanes 11–25) or AZC-treated U-251 MG cells (lanes 6–10). DNA–protein complexes involving specific interactions are shown. Unlabelled competitions were performed using a 25-fold molar excess of cons.HSE (lanes 2 and 7), CLE (lanes 3 and 8), mut.CLE (lanes 4 and 9) or the GRE (glucocorticoid responsive element) (lanes 5 and 10). DNA affinity was analysed using increasing amounts of unlabelled competitors (1-, 2-, 5- and 10-fold molar excess respectively). The unlabelled cons.HSE (lanes 12–15), CLE (lanes 17–20) and mut.CLE (lanes 22–25) oligonucleotides were used as competitors.