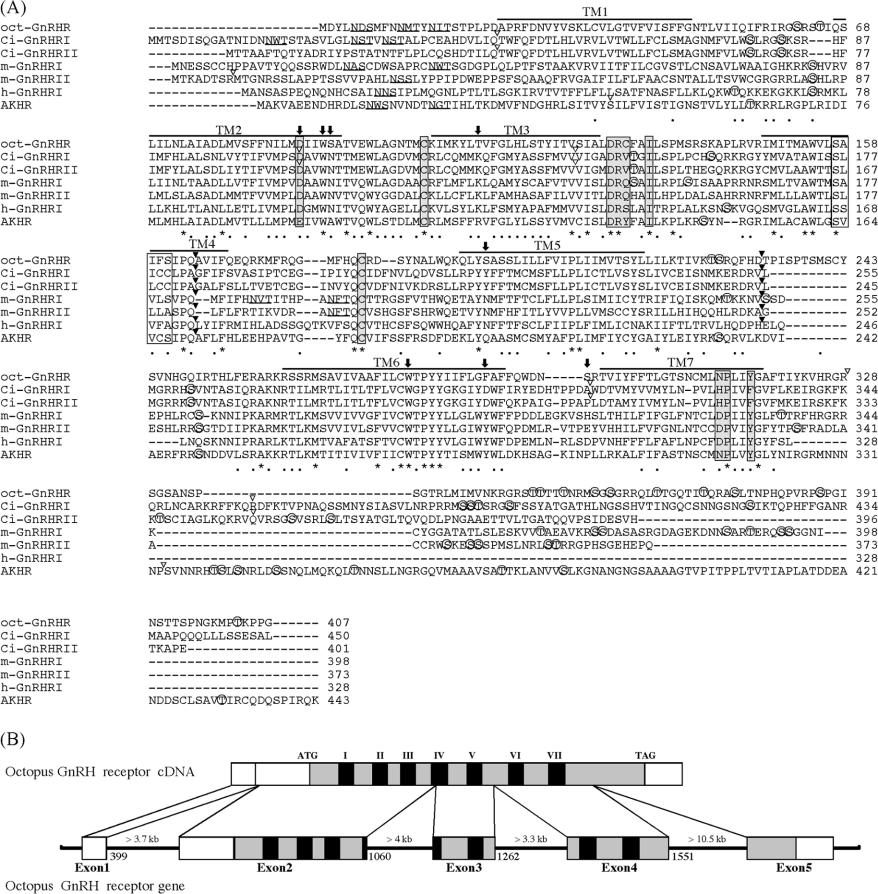
Figure 1. Sequence alignment of oct-GnRHR and other GnRHRs.
(A) The amino acid sequence of oct-GnRHR is aligned with those of the Ciona intestinalis GnRH receptor (Ci-GnRHR I and II [18]), the medaka GnRH receptors (m-GnRHR I and II [19]), human GnRH receptor I (h-GnRHR I [20]) and Drosophila AKH receptor [21]. Amino acid residues conserved in all receptors are indicated by an asterisk. N-linked glycosylation sites are underlined. Bars indicate the seven putative transmembrane domains. Potential phosphorylated serine or threonine residues are marked by open circles. Amino acid residues in grey boxes are believed to play a pivotal role in the GPCR activation, whilst those in white boxes are thought to favour TM helix–helix association [1]. Residues in the agonist-binding pocket described in the text are shown by arrows. The conserved introns in the corresponding genes are indicated the positions by black arrowheads above the sequences; open arrowheads indicate non-conserved introns. (B) Structure organization of the Octopus oct-GnRHR gene. Exons are white boxes, ORFs are grey and TMs are black.