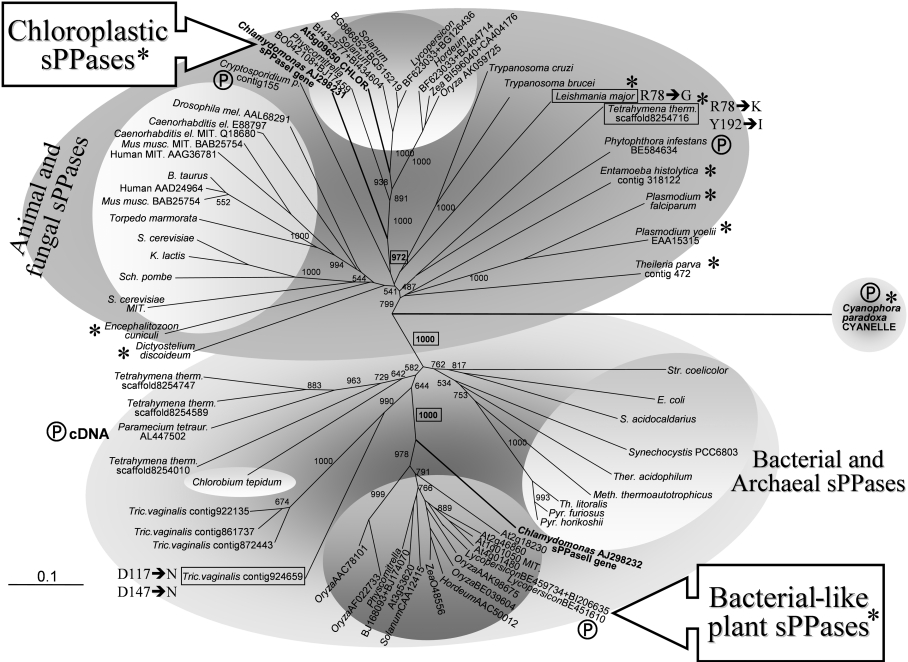
Figure 9. Molecular phylogenetic analysis of the two paralogous family I sPPases identified in photosynthetic eukaryotes and their evolutionary relationships with sPPases of prokaryotes (bacteria and archaea), protists (parasitic and free-living), fungi and metazoa.
Amino acid sequences deduced from algal and plant genes were aligned with sequences of selected family I sPPases using ClustalX. Most sequences have all the amino acid residues reported to be functionally important for sPPase activity [48,49] and the PROSITE motif of family I sPPases. A few sPPase sequences from primitive protists (boxed) exhibit changes in some of these functionally important residues (indicated by Y-sPPase numbering). These sequences are found among natural sPPase mutants, some of them with unusual catalytic features [19,32]. Eukaryotic sequences showing substitutions in residues involved in yeast sPPase homodimer stabilization (Arg51, Trp52 and His87, Y-sPPase numbering; [12]) are marked with asterisks (all chloroplastic sPPases and many orthologous sequences from protists). The few partial sequences used are marked with a Ⓟ symbol. Proteins clearly identified directly, or inferred by computer programs, to have organellar (mitochondrial or chloroplastic) localization, and therefore synthesized as precursors, are labelled accordingly. Numbers at nodes indicate the statistical support (bootstrap values from 1000 replicates) of selected associated groups. The 0.1 bar represents amino acid substitutions per site. Two main clearly defined sPPase assemblies are well supported (bootstrap value 1000, boxed in bold). They cluster on one side as eukaryotic proteins (cytosolic and organellar) from protists, plants, fungi and animals (dark grey upper area), and on the other side prokaryotic (bacterial and archaeal) proteins, as well as bacterial-like sPPases from photosynthetic eukaryotes and some non-photosynthetic protists (ciliates and diplomonads) (light grey lower area). The two clusters of sPPases from photosynthetic eukaryotes (plants and microalgae) tentatively defined in this report, namely chloroplastic sPPase I and bacterial-like sPPase II proteins, are well supported (bootstrap values 972 and 1000 respectively, boxed in bold) and arrange with the eukaryotic (animal and fungal) and prokaryotic sPPases respectively. Accession numbers (in parentheses) for other sequences used are: S. cerevisiae cytosolic sPPase (CAA31629) and mitochondrial sPPase precursor (NP_013994.1); human cytosolic sPPase (AAD24964) and mitochondrial sPPase precursor (AAG36781); Mus musculus cytosolic sPPase (BAB25754) and mitochondrial sPPase precursor (BAB22922); Kluyveromyces lactis cyotosolic sPPase (CAA32466); Encephalitozoon cuniculi (AL590449); Plasmodium falciparum (NP_473275.1); Entamoeba histolytica (contig 318122, TIGR); Dictyostelium discoideum (AU061000 and AU034313); Trypanosoma brucei (contigs 90C7TF and 81B5TF, TIGR); Trypanosoma cruzi (contig TCKGB96TR, TIGR); E. coli (P17288); Synechocystis PCC6803 (AJ252207); Streptomyces coelicolor (O9×819); E. coli (P17288); Sulfolobus acidocaldarius (P50308); Thermoplasma acidophilum (P37981); Methanobacterium thermoautotrophicus (O26363); Thermococcus litoralis (P77992); Pyrococcus horikoshii (O59570). Other putative sPPase sequences obtained with BLAST searches are identified by accession numbers and contig codes indicated in the Figure.