Figure 1.
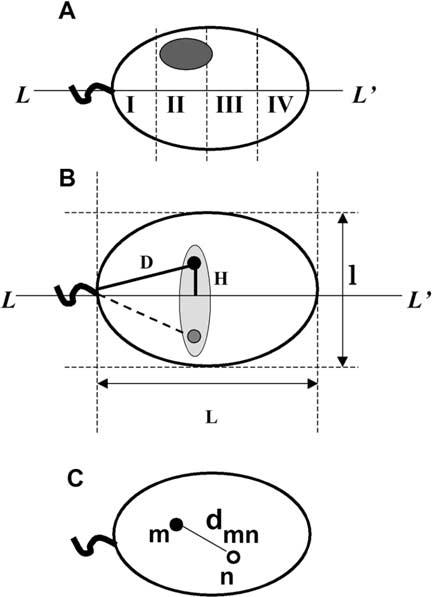
Geometrical parameters of human sperm cell used to determine chromosome positioning. A. Schematic representation of a sperm cell nuclei approximated by an ellipse with the axis of symmetry L-L′. Shaded oval shows an area of hybridization with a chromosome-painting probe. Sperm nucleus is divided into four zones I-IV starting from the basal side that is determined by tail attachment site. B. Parameters, which have been used to describe intranuclear localization of the chromosome-specific centromere hybridization signals. D - distance from the tail attachment point, H - elevation over the long axis. L and l are lengths of the long and short axes. Because of anticipated freedom of rotation along L-L′ we expected random distribution of signals within a schematically shown shaded area. Filled and shaded circles are experimentally observed mirror positions of a centromere. C. Scheme of the sperm nucleus image after multicolor hybridization with two centromere-specific probes. Distances between centers of the FISH signals (dmn) were used to determine relative positioning of centromeres within nuclei.
