Fig. 5.
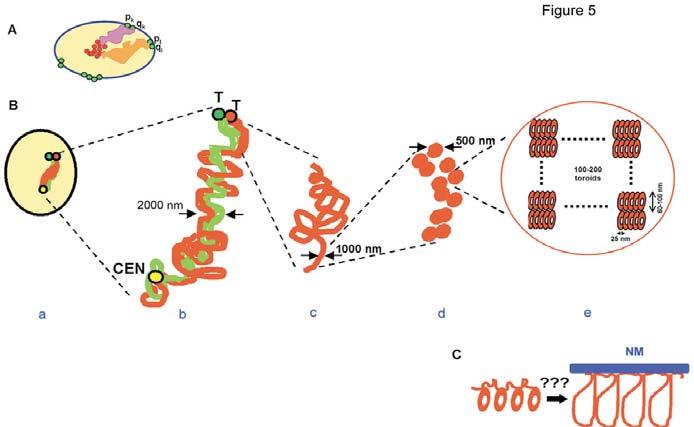
Chromosome architecture in human sperm cells. (A) Model of sperm nuclear architecture proposed on the basis of CEN- and TEL-domain localization (Zalensky, 1988; Zalenskaya and Zalensky, 2000). Only two CTs (orange and pink areas) are shown. They stretch between the sperm chromocenter formed by an association of CENs (red circles) and TELs (green circles), which form dimers at the nuclear periphery. (B) Levels of chromosome architecture in human sperm nuclei, arm-specific chromatins are shown in red and green. (a) Compact CT, p- and q-arms are tightly merged. (b) Extended chromosome hairpin, arms are intertwined or stacked in antiparallel; (c) 1000 nm fiber of an individual arm. (d) Two rows of 500 nm nodular structures that form 1000 nm fiber. (e) Chromatin nodules hypothetically consist of nucleoprotamine toroids. CEN, centromere; T, telomere. (C) Donut-loop model (Ward and Ward, 2004). A thinner chromatin thread, presumably nucleohistone, attached to a nuclear matrix (NM) connects nucleoprotamine toroids. Each toroid consists of a packed 50 kb DNA loop.
