Figure 1.
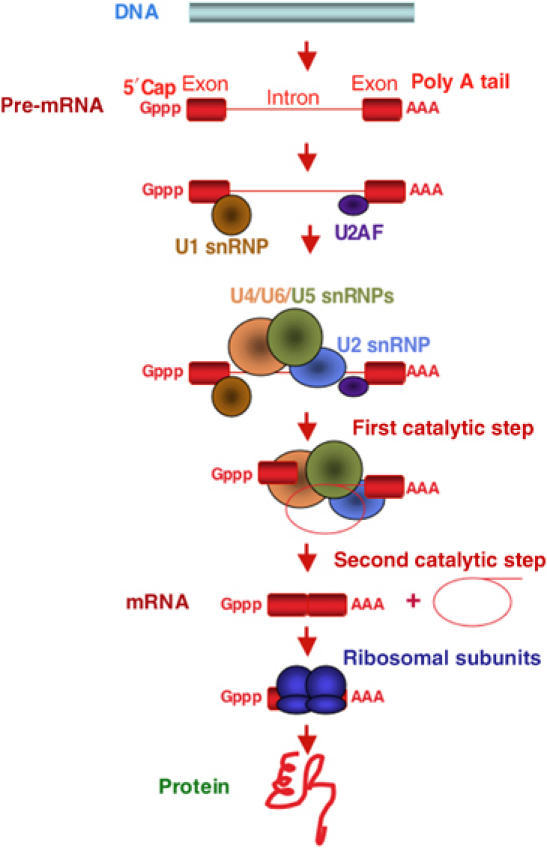
Example of an RNA processing pathway in eukaryotic gene expression. Messenger RNAs are synthesized as precursors containing a 7-methyl-guanidine triphosphate cap at the 5′ end, a polyadenylate tail at the 3′ end and intervening sequences (introns, thin red line) that are eliminated to generate translatable mRNAs where exons (red rectangles) are spliced together. Also represented are some of the components of the spliceosome, the complex that catalyzes intron removal through two consecutive trans-esterification steps. Introns are released in a lariat configuration and are often degraded in the nucleus. Spliceosomal factors include proteins like U2AF and small nuclear RNP particles like U1–U6 snRNPs; many other spliceosomal components are not represented. Most primary transcripts, including rRNAs, tRNAs, snoRNAs and miRNAs, undergo various processing steps catalyzed by protein and/or RNP complexes that eliminate internal or flanking sequences or introduce chemical modifications.
