Figure 6.
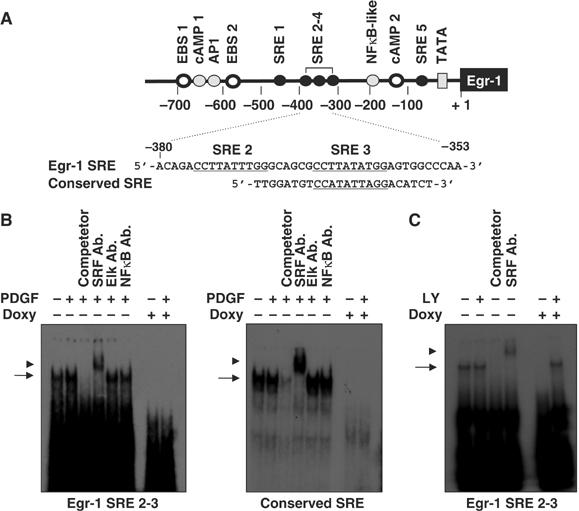
Binding of SRF to the Egr-1 promoter is decreased by the expression of oncogenic H-Ras. (A) Schematic diagram of the oligonucleotide probes used in the EMSA analysis. The SRE sites are underlined. (B) Suppression of the DNA-binding activity of SRF by oncogenic Ras. NIH3T3tet-on/H-RasG12R cells cultured in the absence or presence of doxycycline were serum starved for 24 h in medium that contained 0.5% FBS, and then stimulated with PDGF (50 ng/ml) for 30 min. Nuclear extracts (10 μg/lane) were prepared and subjected to EMSA analysis for the DNA-binding activity of SRF using oligonucleotides derived from the Egr-1 gene (left panel) or conserved SRE gene (right panel). Cold competitor (10-fold molar excess) or the anti-SRF, anti-Elk-1, and anti-NFκB antibodies were incubated for 15 min prior to the addition of 32P-labeled oligonucleotides. The arrow and arrowhead indicate the shift and supershift forms, respectively, corresponding to the position of the SRF protein–DNA complexes. (C) Effect of PI3K inhibition on the DNA-binding activity of SRF. NIH3T3tet-on/H-RasG12R cells were cultured for 24 h, serum starved with 0.5% FBS for an additional 24 h in the absence or presence of doxycycline (2 μg/ml) either with or without LY294002, as indicated. Nuclear extracts (10 μg/lane) were prepared and subjected to EMSA analysis for the DNA-binding activity of SRF, as described in (B).
